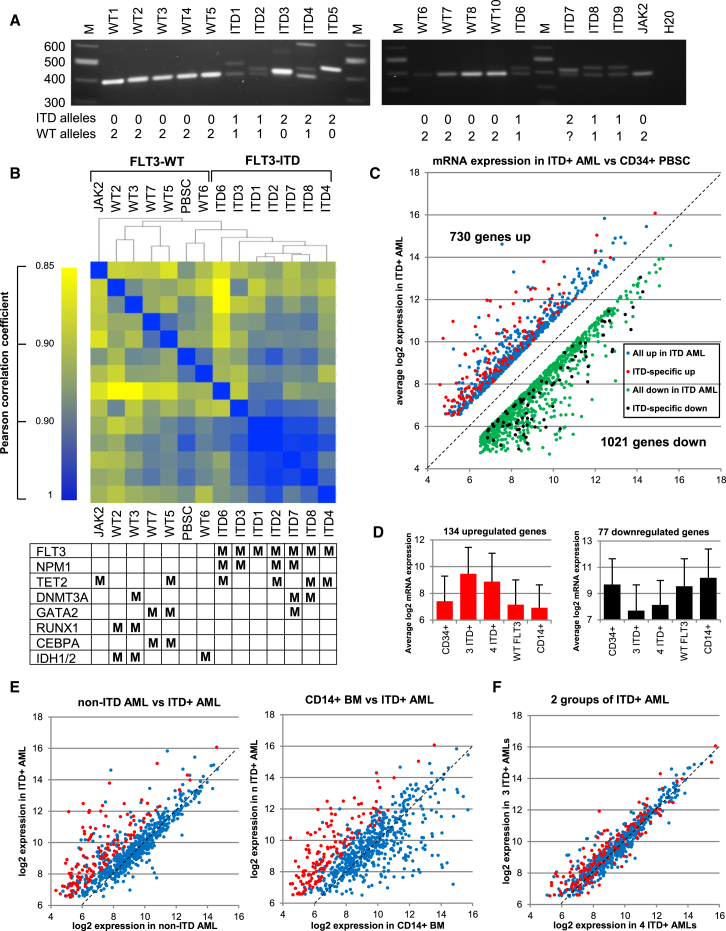
Figure 1.
FLT3-ITD+ AML Displays a Characteristic mRNA Expression Profile
(A) PCR amplification of the transmembrane coding region of the FLT3 gene used to identify ITD mutations. On the left are the sizes of the DNA marker bands (M). Below are the estimated numbers of normal and mutated FLT3 alleles.
(B) Hierarchical clustering of Pearson correlation coefficients of mRNA values. For each sample the mutations present in the most commonly mutated genes are indicated as M in the table underneath.
(C–F) Identification of genes that differ by at least 2-fold in a comparison of the average log2 mRNA microarray values for a core group of three ITD+ AML samples (ITD1, ITD2, and ITD3) to the equivalent values determined from either the average of two independent PBSC samples (C), the average of four WT FLT3 AML samples (WT2, WT3, WT5, and WT7), for CD14+ bone marrow cells (E), or a second group of ITD+ AML samples (ITD4, ITD6, ITD7, and ITD8) used here for validation of the ITD+ pattern (F). The 134 genes that are consistently expressed at 2-fold higher levels in ITD+ AML in each of the three comparisons in (C) and (E) are shown in red, and the remaining genes that are upregulated in at least (C) are shown in blue. In (C) we also highlight 77 genes with values that are consistently 2-fold lower in ITD+ AML compared to each of PBSCs, WT FLT3 AML, and CD14+ cells in black, and the remaining genes that are also downregulated at least in (B) are shown in green. (D) Average log2 mRNA values for the specific groups of 134 upregulated and 77 downregulated genes in each of the 5 groups used in this analysis, with the SD for mRNA expression shown as error bars.