Figure 3.
FLT3-ITD+ AML Has a Characteristic Chromatin Signature
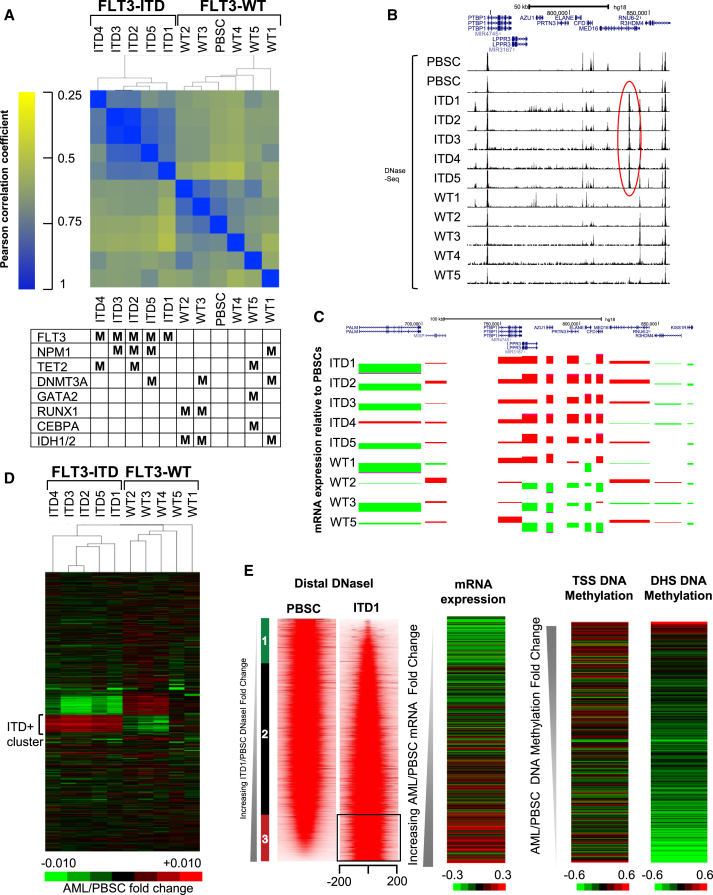
(A) Hierarchical clustering of Pearson correlation coefficients of DHS peaks. For each sample the mutations present in the most commonly mutated genes are indicated as M in the table underneath.
(B and C) UCSC Genome Browser views of the DNase-seq patterns (B), and relative mRNA values (C) of genes surrounding an ITD+ AML-specific DHS in MED16 marked by a red oval, and a cluster of adjacent genes upregulated in ITD+ AML. mRNA values are shown as a ratio of the values detected in the AML samples compared to PBSCs (red: up, green: down).
(D) Heatmap depicting hierarchical clustering of the relative DNase-seq signals seen in each distal DHS peak in each AML sample relative to PBSCs.
(E) Profiles of the DNase-seq signals within each 400-bp window centered on each peak for PBSC and ITD1, with peaks shown in the order of increasing DNase-seq tag count signal for ITD1 relative to PBSC. This analysis includes the union of all peaks present in either ITD1 or in PBSC. Shown to the right of the DNase-seq profiles are the relative mRNA expression values for genes with the nearest transcription start sites (TSS) in ITD1 relative to PBSC, and the DNA methylation signals for the nearest TSS and for the DHS in ITD1 relative to PBSCs.