Figure 7.
ITD-Specific DHS Motifs Are Occupied in ITD+ AML
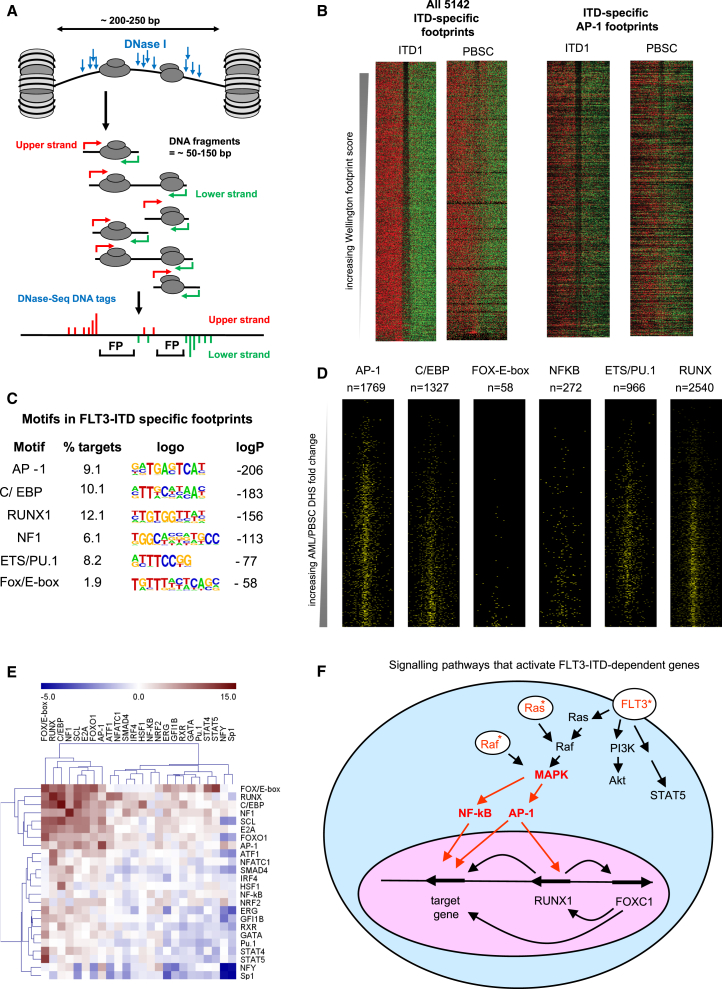
(A) Model depicting the generation of DNase I footprints at DHSs.
(B) DNase I cleavage patterns within ITD1-specific footprints predicted by Wellington. Upper strand cut sites are shown in red and lower strand cut sites in green within a 200-bp window centered on each footprint (gap), for all 5,142 ITD-specific footprints, and for those containing AP-1 sites.
(C) Analysis of overrepresented binding motifs within each footprint using HOMER.
(D) Profiles of motifs in ITD-specific footprints plotted on the same DHS axes as used in Figure 2E.
(E) Analysis of statistically significant co-localization of the indicated footprinted motifs within 50 bp of each other using bootstrapping analysis (Z score).
(F) Model of an ITD-specific transcriptional network based on our data.