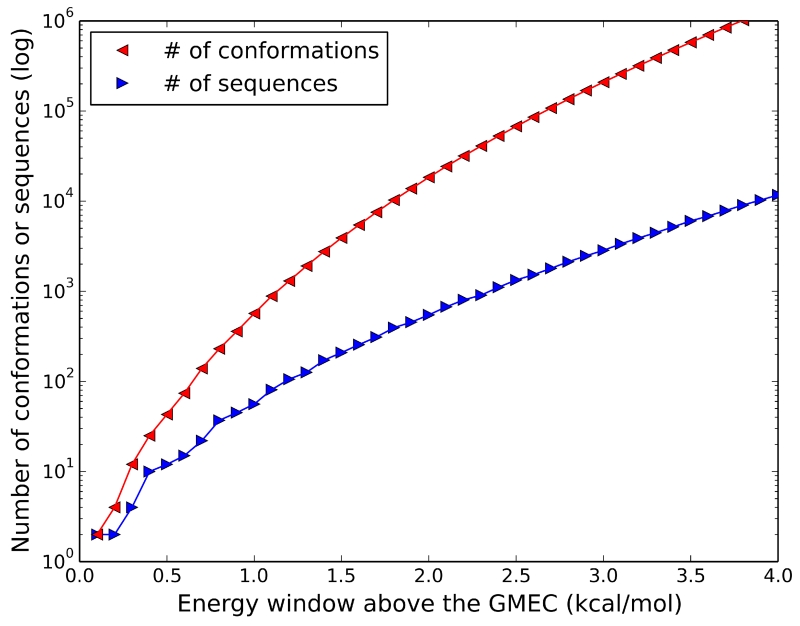
Figure 7. The new Sequence-A* algorithm enumerates sequences much faster than Conformation-A*.
To experimentally test protein design predictions, it is often beneficial to predict many low-energy sequences rather than the single GMEC. Conformation-A* methods enumerate conformations in a gap-free, in-order ranking of low-energy conformations. However, most of the low-energy conformations often belong to a small set of protein sequences. Consequently, Conformation-A* enumerates many more conformations than sequences. The number of unique conformations (red) and sequences (blue) are shown for the protein core design of toxin II (PDB id: 1AHO). Each plotted data point shows the number of unique sequences (or conformations) within the given energy cutoff of the GMEC’s energy. Due to the explosion of conformations within 4.0 kcal/mol of the GMEC’s energy, Conformation-A* was unable to find all unique sequences within 4.0 kcal/mol of the GMEC’s energy within seven days. In contrast, by directly enumerating sequences, Sequence-A* was able to find all unique sequences in 62 minutes.