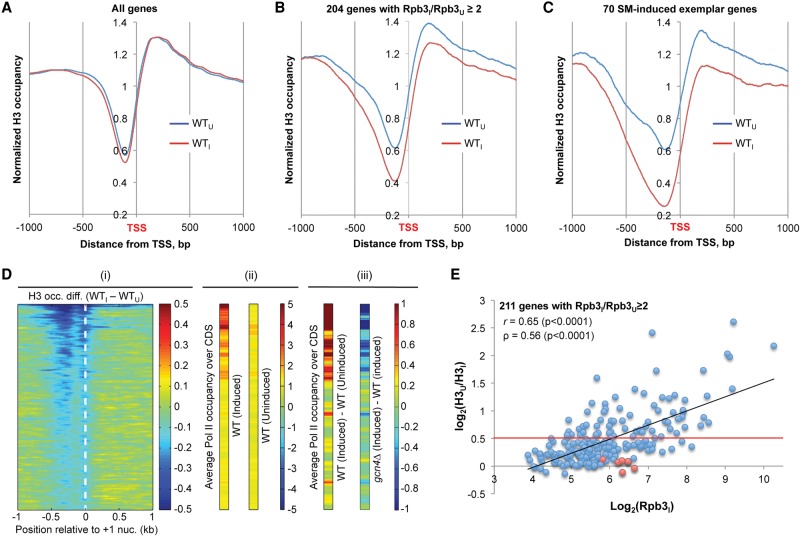
Figure 2.
Eviction of H3 from promoters correlates with induction of Pol II occupancies at SM-induced genes. (A–C) Plots of average H3 occupancies at each base pair aligned at the TSS, calculated from ChIP-seq data combined from three biological replicates. The relative H3 occupancy at each base pair surrounding the TSS, normalized to the average occupancy on the respective chromosome, was averaged for the indicated gene sets, and the resulting values are plotted for the interval spanning 1000 bp 5′ to 1000 bp 3′ of the TSS. Blue indicates WTU; red, WTI. (D) Heat map depictions of changes in relative H3 and Rpb3 occupancies for 1000 genes with the largest reductions in H3 promoter occupancies on induction of WT cells. (i) Relative H3 occupancy differences between WTI and WTU cells in the region [−1000, +1000] around the +1 nucleosome dyad, sorted in descending order, and color-coded as shown on right. (ii) Relative Rpb3 occupancies, averaged over CDS, in WTI and WTU cells for the same gene order, color-coded as shown on right. (iii) Differences in relative Rpb3 occupancies between WTI and WTU cells (left) or between gcn4Δ and WT cells (right), color-coded as on far right. ChIP-seq data for H3 and Rpb3 were combined from three biological replicates. (E) Scatterplot of Rpb3 occupancies per base pair in CDS in WTI cells versus the ratio of average relative H3 occupancies in the [−1,NDR,+1] intervals of WTU versus WTI cells for 211 SM-induced genes, calculated from ChIP-seq data from three biological replicates. Pearson's (r) and Spearman's (ρ) correlation coefficients are indicated. Seventy-three genes above the red line exhibit a reduction in H3 occupancy of ≥30% on SM-induction. For genes in red exhibiting high-level Rbp3I with little or no H3 eviction (SNZ1, FMN1, PHM8, ARG8, HSP78, MCH1), nucleosome sliding to a new location in the promoter, or eviction of only H2A:H2B dimers, may suffice for PIC assembly.