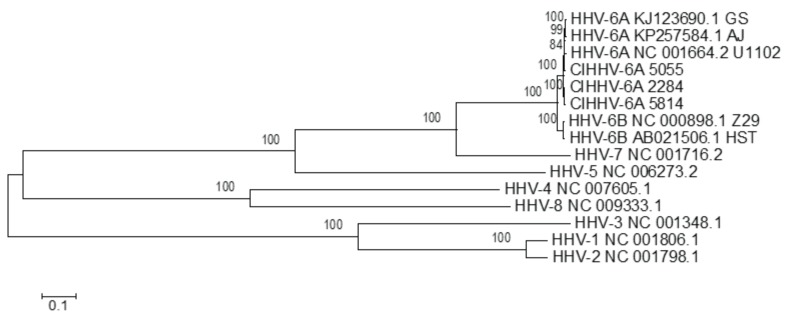
Figure 1.
Phylogenetic analyses of CiHHV-6A compared to human herpesvirus representative of herpesviridae species in alpha, beta, and gamma herpesvirus sub-families. Maximum likelihood method was used in Mega 5.1 using core conserved genes to compare the relationship of CiHHV-6A 5055, 2284, and 5814 genomes to betaherpesviruses including the Roseolovirus genera of HHV-6A, HHV-6B, HHV-7, together with Human cytomegalovirus (HHV-5); gammaherpesviruses Epstein Barr virus (HHV-4) and Kaposi’s sarcoma associated herpesvirus (HHV-8); alphaherpesviruses Herpes Simplex Virus (HSV) type 1, HSV-1 (HHV-1), and HSV-2 (HHV-2). The reference sequences accession numbers used are shown in the figure. Core genes included homologues of capsid triplex subunit 1 (U29), small capsid protein (U32), large tegument protein (U3), large tegument binding protein (U30), cytoplasmic egress tegument protein (U71), cytoplasmic egress facilitator-1b (U44), glycoproteins gB, gL, gM (U39, U82, U72), multifunctional expression regulator (U42), DNA polymerase catalytic subunit (U38), DNA polymerase processivity subunit (U27), helicase-primase RNA polymerase subunit (U43), helicase primase subunit (U74), single-stranded DNA-binding protein (U41), alkaline deoxyribonuclease (U0), uracil DNA glycolase (U81), ribonucleotide reductase large subunit (U28), capsid transport nuclear protein (U36), DNA packaging terminase subunit 2 (U40), terminase binding protein (U35), nuclear egress membrane protein (U34), and nuclear egress lamina protein (U37). Bootstrapping (1000) shows percentage of trees where taxa cluster together and scale shows branch lengths measured in number of substitutions per site.