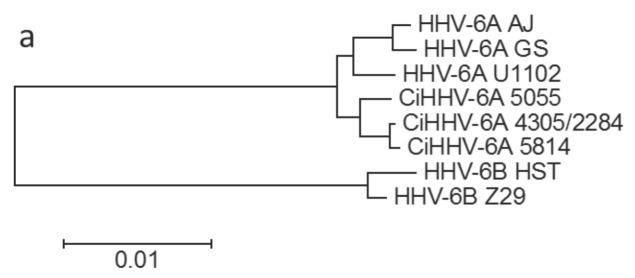
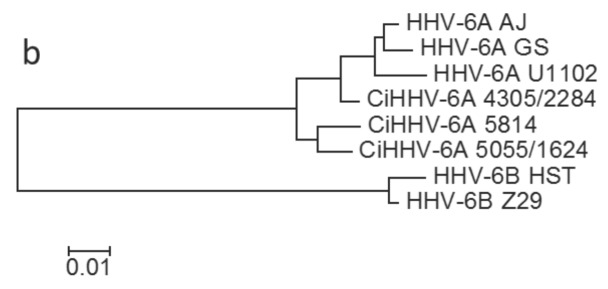
Figure 2.
Phylogenetic tree analyses of CiHHV-6A and Herpesviridae, conserved core and variable genes. Maximum likelihood method was used in Mega 5.1 using Bestfit model of General Time Reversible with gamma distribution (methods) with maximized amino acid alignment followed by back-translation and phylogenetic reconstruction. (a) Conserved core genes were compared showing relationships. These included 14 genes encoding mainly HHV-6A/B conserved structural and DNA replication components and included U31-39, U42-44, U74, U81-82; (b) Variable genes as defined in Table 1, included 16 genes and were compared to homologues in genomes of strains of HHV-6A/B virus. Bootstrapping (1000) shows percentage of trees where taxa cluster together were >95% for all nodes. The scales shows branch lengths measured in number of substitutions per site. HHV-6B was used as an out-group to analyze relationships between HHV-6A and CiHHV-6A.