Fig. 2.
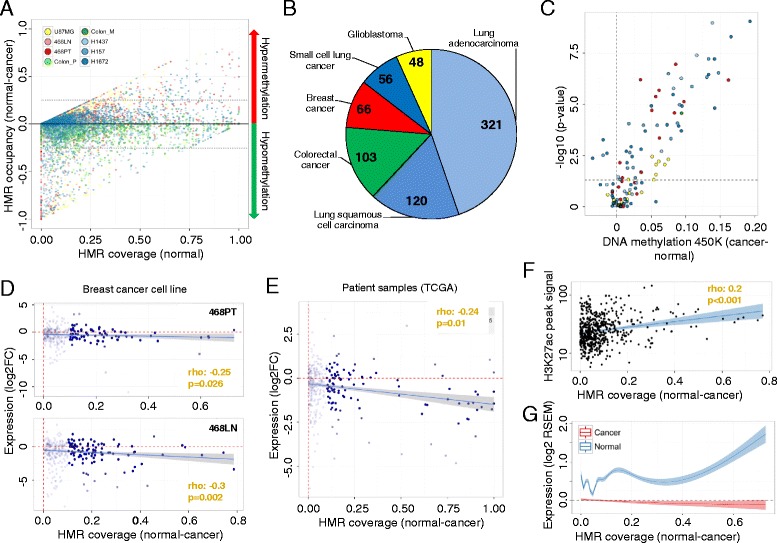
Cancer-specific alterations in DNA methylation within super-enhancer regions determined using WGBS. a Difference in DNA methylation levels (occupancy of hypomethylated regions (HMRs)) between cancer (n = 8) and normal (n = 5) samples paired within their respective tissue contexts (y-axis). HMR occupancy of normal tissues is indicated (x-axis) and cancer sample types are color-coded and the threshold indicated (dotted line; δ HMR occupancy 25 %). b Sample distribution of 714 cancer samples analyzed on the HumanMethylation450 BeadChip. c Validation of DNA hypermethylation at super-enhancers in 714 cancer samples using the HumanMethylation450 BeadChip (450 K). Significance was assessed by differential DNA methylation levels and the Student’s t-test (p value), comparing normal and cancer samples and averaging over the analyzed CpG (≥3) within a super-enhancer region (FDR < 0.05). The cancer samples are color-coded as defined in (b). d The association between HMR occupancy (WGBS) and target gene expression (RNA-seq) is assessed comparing normal breast (MCF10A) and the primary (468PT, upper panel) and metastatic (468LN, lower panel) breast cancer cell lines. Expression data are displayed as log transformed fold-change (log2FC) and significances of a Spearman’s correlation test are indicated. e Differences in HMR occupancy (WGBS) and target gene expression (RNA-seq, scaled log expression) are displayed comparing matched normal breast and primary carcinoma samples (TCGA [16], n = 25). f Association of H3K27ac signal (ChIP-seq) and differential HMR occupancy (WGBS) at hypermethylated super-enhancers. H3K27ac signals were retrieved from normal breast tissue [11]. g Smoothed (GAM) scaled log expression values of super-enhancer-related genes in matched normal and cancer samples (TCGA [16], n = 25) plotted against the difference in HMR occupancy (WGBS) for all super-enhancers gaining methylation in cancer. GAM Generalized Additive Model, RSEM RNA-Sequencing by Expectation Maximization
