Fig. 1.
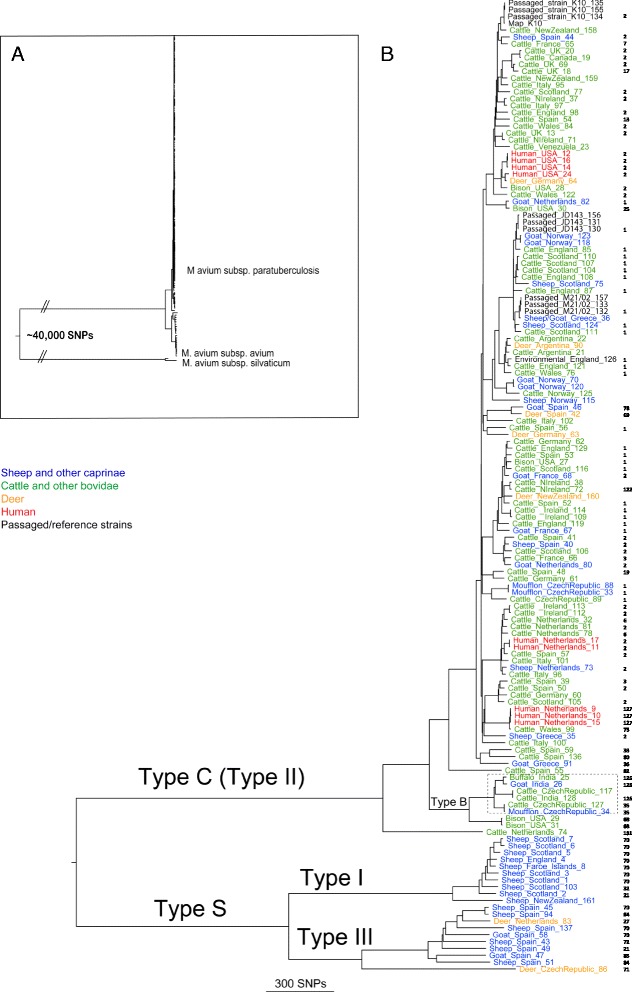
Whole genome SNP-based phylogenetic tree of strains included in this study. a Phylogenetic tree of Map and non-Map strains built using FastTree [52]. Map strains form a single clade and are separated from other mycobacterial species by at least 40,000 SNPs (branches shortened for illustrative purposes). b Maximum likelihood phylogenetic tree of Map strains sequenced as part of this study. The tree was created using RAxML [30], and is based on SNPs identified through mapping to Map-K10 as described in the text. Branches are annotated with the host, country of origin and isolate MAPMRI numbers. Numbers in black to the far right represent the INMV profiles. Previously described lineages are labeled. The dashed box represents strains designated as the Indian bison type. Bootstrap values are shown in Additional file 2: Figure S1
