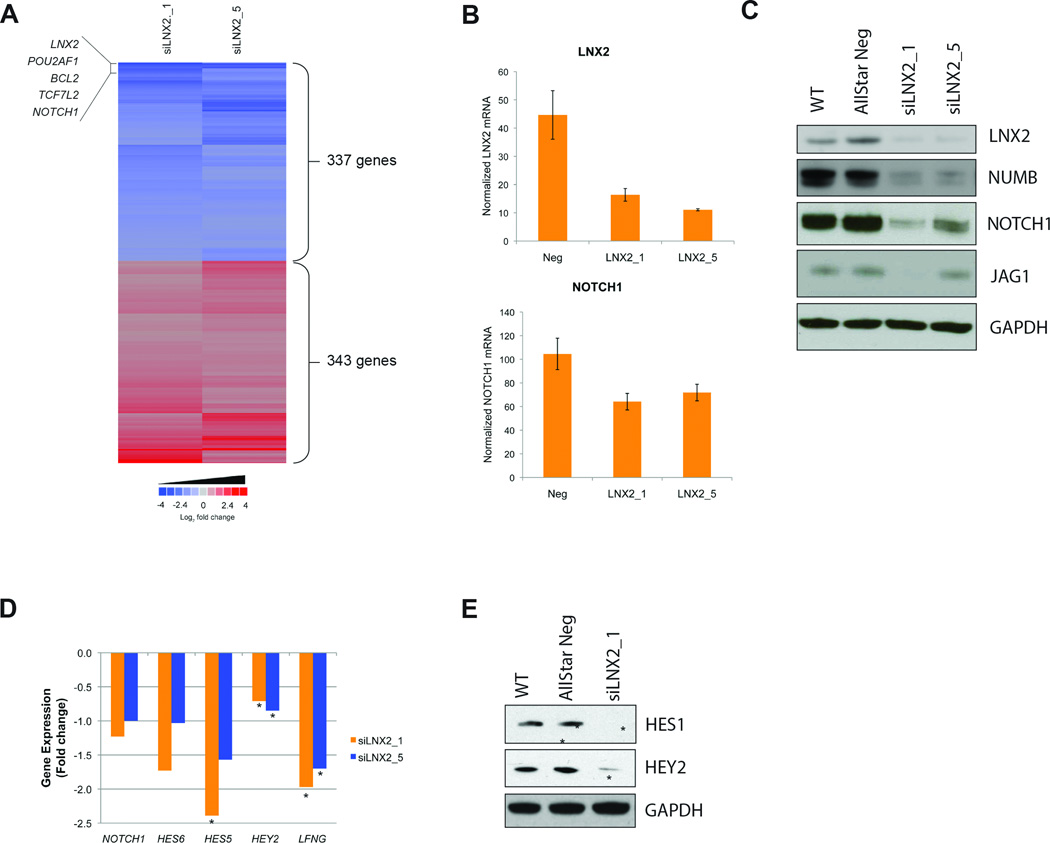
Figure 5. Analysis of the RNAi signature of the candidate gene LNX2.
(A) Heatmap of the commonly deregulated genes upon silencing of LNX2 in SW480. Six hundred-eighty genes were significantly deregulated in the LNX2-specific RNAi signature. Note that two different siRNAs against this gene result in a very reproducible deregulation of the same signature genes. Among the most downregulated genes (blue) are the transcription factors NOTCH1 and TCF7L2, and the anti-apoptotic gene BCL2. (B) qRT-PCR for LNX2 and NOTCH1 expression levels after silencing of LNX2. The successful silencing of LNX2 correlates with reduced expression of NOTCH1, confirming the expression levels measured on the arrays. (C) Western Blot analysis for the confirmation of silencing on the protein level for LNX2, NOTCH1, NUMB, and JAG1 after silencing LNX2 with two independent siRNAs. Controls: untransfected cells (WT) and cells transfected with AllStarNeg. The results confirm the gene expression data. (D) Gene expression changes of NOTCH1 and the NOTCH downstream targets HES6, HES5, HEY2, and LFGN based on microarray data after silencing LNX2 with two independent siRNAs (* P<0.05). (E) Western Blot analysis demonstrating significant downregulation of NOTCH regulated proteins HES1 and HEY2, indicating negative regulation of NOTCH signaling after silencing LNX2, in accordance with the gene expression data presented in (D).