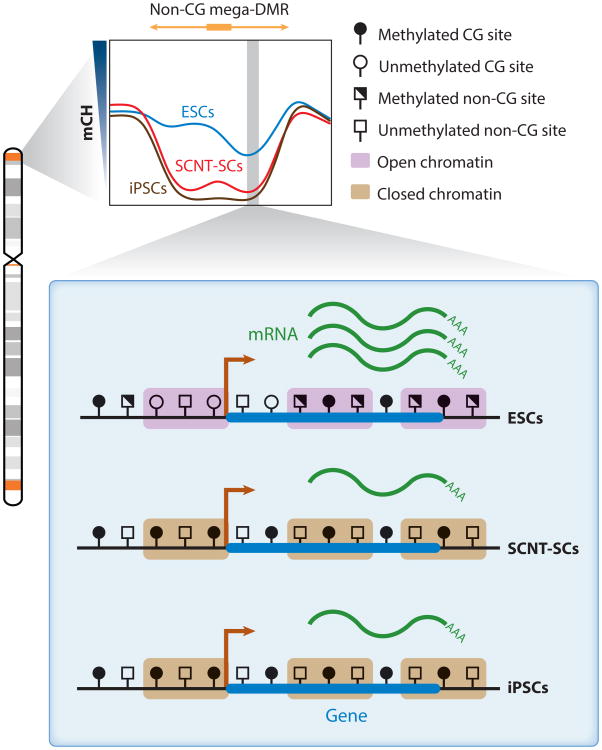
Figure 4.
Non-CG mega-DMRs in ESCs, SCNT-SCs, and iPSCs (57, 61). The non-CG mega-DMRs are shown in orange in the chromosome diagram and tend to be proximal to telomeres and centromeres (64, 76); the gray shades are cytogenetic bands. The vast majority of non-CG mega-DMRs are hypomethylated in SCNT-SCs and iPSCs compared with ESCs (shown in the mCH profile of non-CG mega-DMRs). In SCNT-SCs and iPSCs, the heterochromatin mark H3K9me3 is enriched in these regions. In addition, the transcription of genes within non-CG mega-DMRs is disrupted, and the promoters of these genes are typically CG hypermethylated in SCNT-SCs and iPSCs compared with ESCs. Abbreviations: ESC, embryonic stem cell; H3K9me3, histone H3 with trimethylated lysine 9; iPSC, induced pluripotent stem cell; mCH, non-CG methylation; mega-DMR, megabase-scale differentially methylated region; SCNT-SC, somatic cell nuclear transfer stem cell.