Figure 5.
DNA Methylation Differences Are Associated with DE Genes
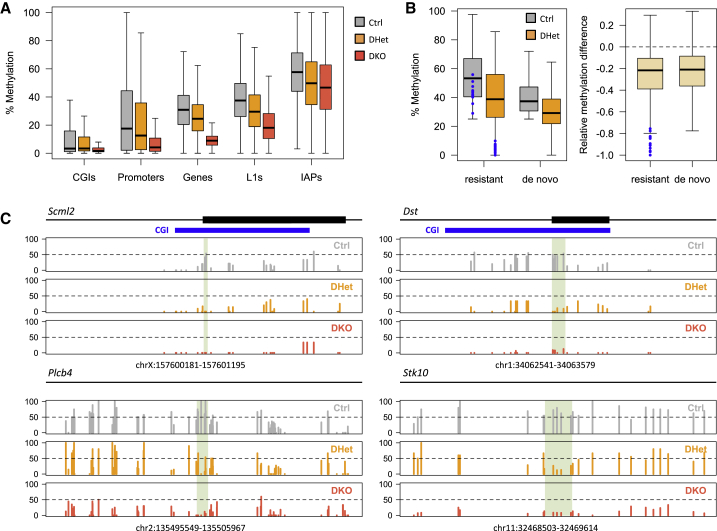
(A) DNA methylation profiling by BS-seq shows that DHet EPCs have a slight genome-wide reduction of DNA methylation.
(B) CGIs methylated (>25%) in Ctrl EPCs were separated into reprogramming-resistant (>25% methylation in blastocyst; Kobayashi et al., 2012) or de novo methylated (<15% methylation in blastocyst). Methylation levels in Ctrl and DHet EPCs (left) and the relative methylation change between the two (right) shows that both subsets are hypomethylated to a similar extent in DHet EPCs; maternally methylated ICRs (blue) undergo more extensive methylation loss.
(C) BS-seq profiles of methylation levels at example loci containing DHet DMRs (highlighted in green) that are associated with genes displaying altered gene expression.
See also Figure S5.