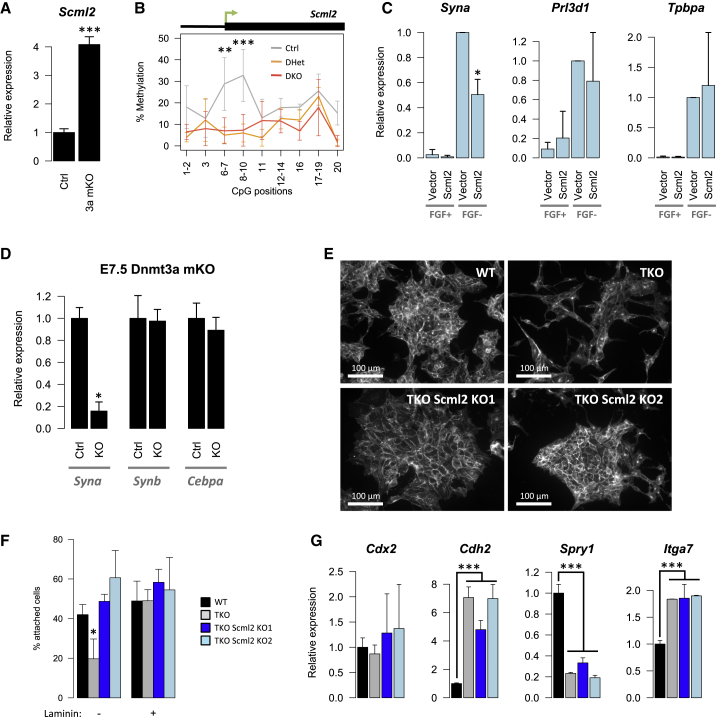
Figure 6.
Scml2 Is Controlled by DNA Methylation and Affects SynT Formation and Cell Adhesion
(A) RT-qPCR of Dnmt3a mKO EPCs confirms that Scml2 is controlled by oocyte methylation.
(B) Methylation analysis by Sequenom MassARRAY in E7.5 male EPCs, confirming the DMR at an intragenic TSS of Scml2. Each data point may include more than one CpG from the amplicon, as indicated on the x axis.
(C) RT-qPCR analysis of TSCs grown in FGF+ (TSC conditions) or FGF− (differentiation conditions) medium for 6 days, with or without Scml2 overexpression.
(D) Expression of Syna is reduced in Dnmt3a mKO EPCs, whereas markers of SynT-II Synb and Cebpa are unaffected.
(E) E-cadherin staining of two independent Scml2 knockout clones from TKO TSCs shows a rescue of the morphological alterations seen in TKO TSCs.
(F) Scml2 KO on TKO TSCs also rescues the defect in cell adhesion to cell culture wells in the absence of laminin.
(G) RT-qPCR analysis of TKO Scml2 KO clones shows maintained expression of the TSC marker Cdx2; the expression of genes involved in cell adhesion is not rescued upon Scml2 deletion.
Error bars represent SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; t test comparing WT and Dnmt3a mKO EPCs (A and D) or Scml2-expressing TSCs versus vector control (C), or ANOVA with post hoc tests comparing Ctrl with DHet/DKO (B) or TKO TSC lines with WT TSCs. See also Figure S6.