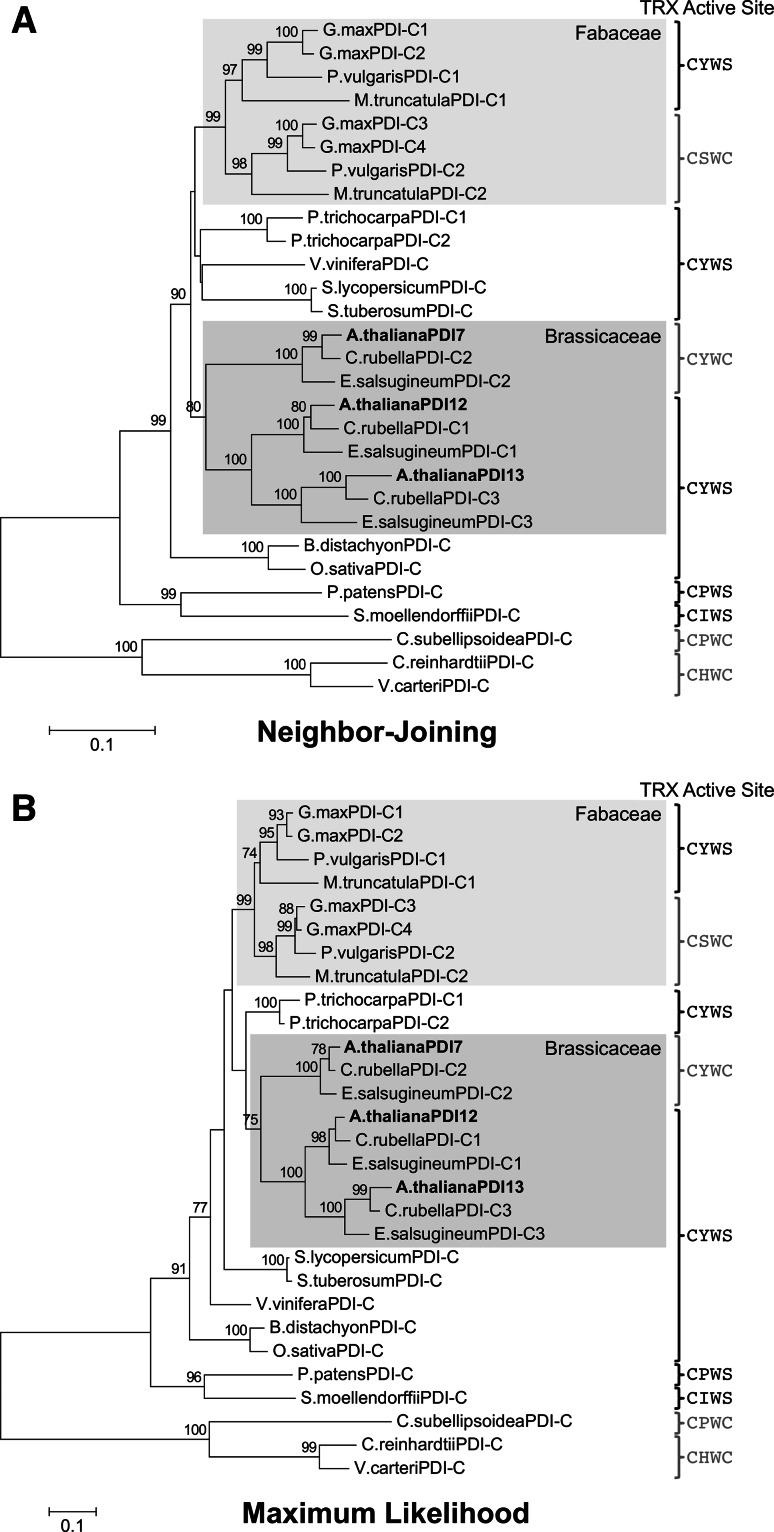
Fig. 3.
Phylogenetic analysis of plant PDI-C isoforms. a The unrooted NJ tree was generated with evolutionary distances computed using the Poisson correction method. The tree is drawn to scale, with branch lengths proportional to the number of amino acid substitutions per site. b The unrooted ML tree was generated using the LG + G model, with 5 discrete gamma categories (+G, parameter = 0.9871). The tree with the highest log likelihood (−7366.6878) is shown, with branch lengths measured in the number of substitutions per site. NJ and ML analyses were performed using a Gblocks-trimmed multiple sequence alignment consisting of 407 positions. Support values are shown above the branches, and are calculated from 1000 bootstrap replicates. Only bootstrap values ≥70 % are shown. The Fabaceae and Brassicaceae clades are shaded. The thioredoxin (TRX) domain active site sequence motifs of the various PDI-C isoforms are shown on the right, with motifs containing two Cys residues highlighted in red (colour figure online)