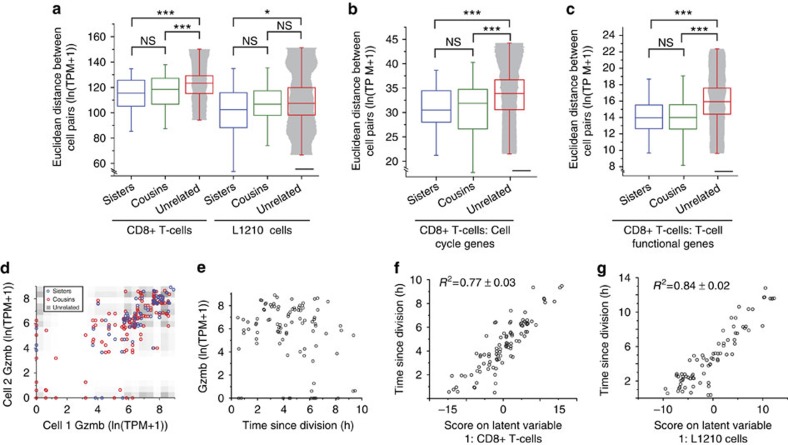
Figure 2. Single-cell RNA-seq with known lineage relationships and times since division.
(a) Comparison of Euclidean distances (measured in log-transformed transcripts per million) between sister cells, cousin cells and unrelated cells for CD8+ T-cells (n=43, 73 and 4,544, respectively) and L1210 cells (n=37, 60 and 3,064, respectively) for the entire gene set (9,997 genes and 10,658 genes for CD8+ T-cells and L1210, respectively). Groups were compared with a Mann–Whitney U-test (Methods). The shaded area overlay on the unrelated cell pair measurements has a width corresponding to differences in time since division for these cells. The widths were constructed using a 250-point moving average of the pairwise differences in time since division for visual clarity. Scale bar, 3 h. (b) Same analysis as in (a) applied to a subset of genes with cell cycle-related gene annotations (688 genes total) for CD8+ T-cells. (c) Same analysis as in a,b applied to a subset of genes with gene annotations related to T-cell activation and function (142 genes total) for CD8+ T-cells. (Groups were compared with a Mann–Whitney U-test. After Bonferroni correction: *P<0.05, **P<0.01, ***P<0.001. Not-significant (NS) indicates a P value >0.05.) (d) Plot of Granzyme B expression levels (measured in log-transformed transcripts per million) in sister cell pairs (blue circles), cousin cell pairs (red circles) and unrelated cell pairs (grey scale density plot with darkest regions corresponding to highest relative occupancy). The correlation coefficients of related and unrelated cell pairs were compared with a Fisher's z transformation (P=0.005). (e) Plot of Granzyme B expression levels as a function of the time since division for each single cell. (f,g) Plot of scores on the first latent variable of partial least squares regression models constructed with expression measurements as predictor variables and the time since division for each single cell as the response variable for CD8+ T-cells (f) and L1210 cells (g). The final models were constructed with genes corresponding to the top 300 VIP scores for each cell type (Supplementary Fig. 9, Methods).