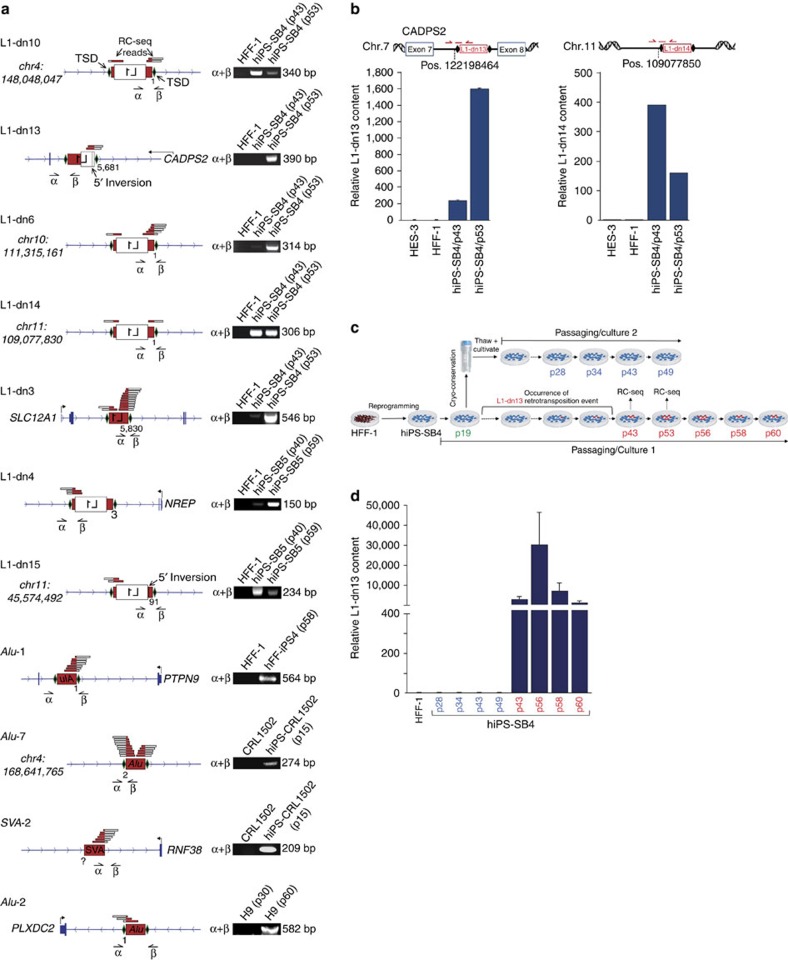
Figure 3. RC-seq reveals endogenous de novo L1, Alu and SVA retrotransposition in pluripotent stem cells.
(a) Structures of validated de novo L1, Alu and SVA retrotransposition events (red box, untranslated region; white box, L1 ORF; green diamonds, TSDs). Names of insertions (for example, L1-dn10), and gene (for example, SLC12A1) or chromosomal positions for intergenic insertions are listed. RC-seq reads are aligned above the insertions (red/white bars). Nucleotide positions at 5′ ends of L1 and Alu insertions refer to L1.3 and AluYb8 reference sequences, respectively. Corresponding validation PCRs are presented on the right. α and β, validation primers. (b) Relative L1-dn13 and L1-dn14 copy numbers at hiPS-SB4 passages 43 and 53 were determined by qPCR. Binding sites of the TaqMan primer/probe combinations specific for the 5′ junctions of insertions L1-dn13 or L1-dn14 are shown (Top panels, red arrows and lines). Genomic DNAs from parental HFF-1 cells, and HES-3 cells served as negative controls. For normalization, a primer/probe combination specific for the human single-copy gene RPP25 was used. ΔΔCt values measured the relative L1-dn13 and L1-dn14 insertion content, respectively, normalized to the parental cell line HFF-1. Bars, arithmetic means±s.e.m. of technical triplicates. Due to the minimal s.e.m. observed in the L1-dn14-specific qPCR (right panel), error bars are not visible. (c) Passaging scheme of the hiPS-SB4 line harbouring L1-dn13. After reprogramming of HFF-1 cells into the hiPS-SB4 line, hiPSCs were cultivated for 60 passages (culture 1). Genomic DNA (gDNA) was isolated from culture 1 at passages shown in red. Cells of passage 19 were split and half of the culture was cryo-preserved and cultivated again after several weeks of cryo-preservation (culture 2). gDNA was isolated from passages shown in blue. (d) Relative L1-dn13 content at passages 43, 56, 58 and 60 of culture 1 (red lettering) and at passages 28, 34, 43 and 49 of culture 2 (blue lettering) were quantified by qPCR. L1-dn13 is present in passages 43 to 60 of culture 1, but absent from culture 2.