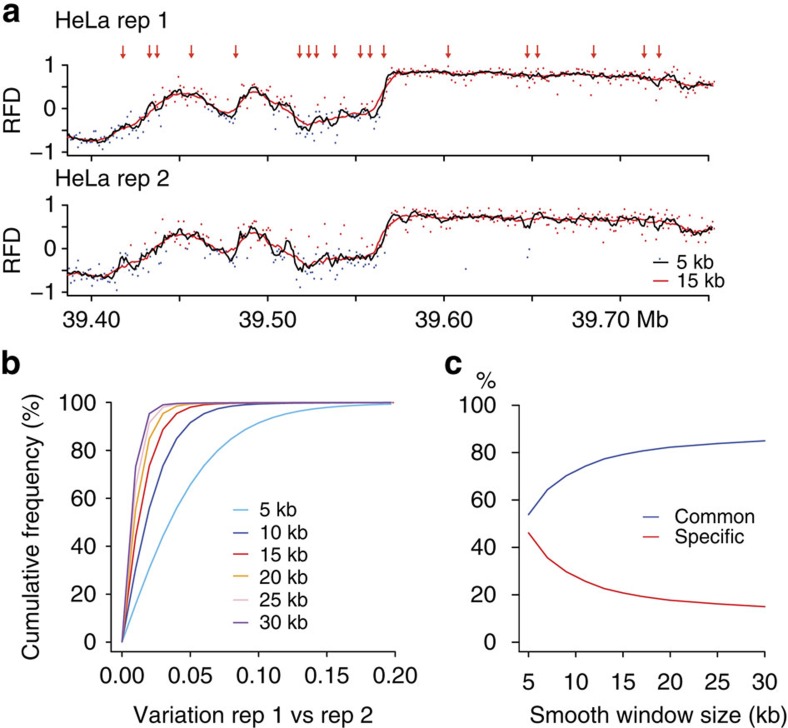
Figure 4. Effect of HMM sliding window on AS detection reproducibility between biological replicates.
(a) RFD profiles corresponding to two OK-seq experiments (rep 1 and rep 2); RFD was computed in sliding windows of size l=5 kb (dark line) and l=15 kb (red line); at each point n of the RFD profiles, the slope was computed as s(n)=RFDn+1−RFDn ; the difference ∂(n)=s1(n)−s2(n) between the slopes of the two profiles was computed along the genome; red arrows indicate the positions at which ∂(n)>0.10 in the case where l=5 kb. (b) ∂(n) was computed for various values of l (l=5, 10, 15, 20, 25, 30 kb); the cumulated proportion of ∂(n) (proportion of positions for which ∂(n)>x) was displayed for each l value. For l=5 kb, 40% of genome positions presented slopes differing by more than 0.05 (light blue). This proportion dropped down to 10 % for l=10 kb, and to 4% for l=15 kb. This showed that the difference between the two profiles strongly decreased for l ≥15 kb. (c), AS of the rep 1 and rep 2 profiles were determined using the HMM procedure (see Methods section) for various sizes of l. The proportion of AS common to both replicate profiles (blue) or specific to one of the two profiles (red) was displayed as a function of l. The proportion of AS specific to only one replicate, that is, that would not be considered as bona fide AS, decreased from almost 50% for l=5 kb to 20% for l=15kb and then decreased more slowly for larger l values. This means that analysis of RFD profiles with l<15 kb would lead to an excessive proportion of AS not conserved between replicates, likely corresponding to too small signal/noise values.