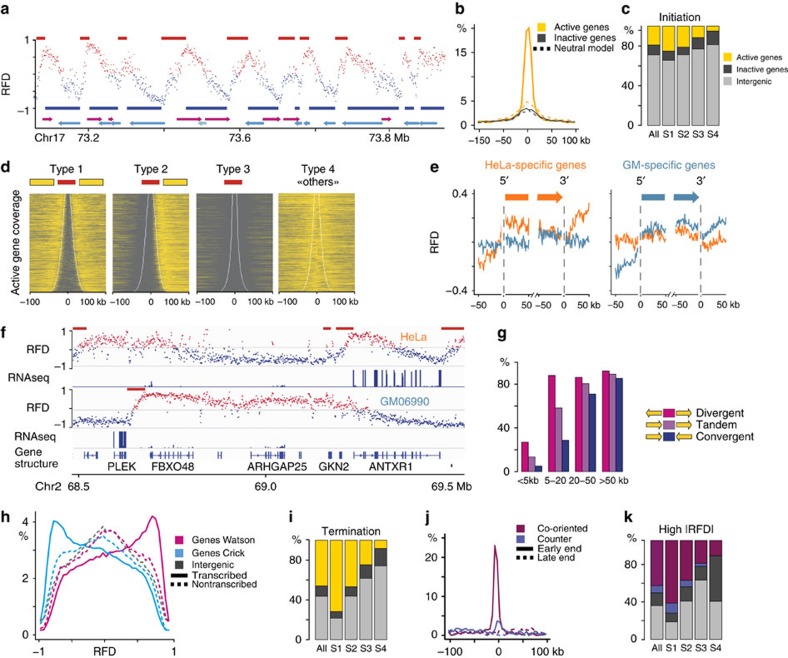
Figure 5. Transcription impact on replication landscape.
(a) Red (blue) lines above (below) HeLa RFD profile indicate ascending (descending) HMM-detected segments (see Methods section); magenta and cyan arrows indicate genes. (b) Line, histogram of distance between an AS end and the nearest active (yellow) or inactive (charcoal) gene; dots, neutral model with randomly positioned AS (see Methods section). (c) Percentage of AS covering (majority rule) active genes (yellow), inactive genes (charcoal), intergenic regions (silver), shown for all AS or for AS in successive periods of S phase (S1–S4; see Methods section). (d) Heatmap of active genes around the four types of HeLa AS described in the text; yellow boxes, active genes flanking AS (red lines). (e) Mean RFD profiles of HeLa (orange) and GM06990 (blue) around 5′ and 3′ ends of isolated genes (see Methods section) expressed specifically in HeLa (left) or GM06990 (right). (f) Examples of AS associated with genes specifically expressed in HeLa (ANTXR1) or GM06990 (PLEK). (g) Proportions of segments between active genes, which overlap (≥1 nt) an AS, for different distances and orientations of flanking genes (HeLa). (h), histograms of HeLa RFD for intergenes (grey), genes oriented rightward (magenta) or leftward (cyan), transcribed (lines) or not (dash). (i) Percentage of HeLa DS covering genes or not, displayed as in c. (j) Histogram of distance between early- (line) or late- (dash) replicating end of high |RFD| (|RFD|>0.6) FS and 5′ or 3′ end of the nearest co-oriented (eggplant) or counter-oriented (blue) active gene. (k) Percentage of HeLa high |RFD| FS covered (majority rule) by sequences from co-oriented (eggplant) or counter-oriented (blue) active genes, inactive genes (charcoal), intergenic regions (silver).