Abstract
Circular RNA (circRNA) is a type of RNAs which, unlike the better known linear RNA, forms a covalently closed continuous loop. They have emerged recently as a new player in governing fundamental biological processes. However it remains elusive about the correlation of hsa_circ_001988 abundance with colorectal cancer. To investigate the circular RNA expression in colorectal cancer, the targeted hsa_circ_001988 was selected from next generation sequence data base generated in house and then designed divergent primers to amplify hsa_circ_001988 and sequenced it for validation. The expression of hsa_circ_001988 in 31 matched colorectal cancer tissue and normal colon mucosa was analyzed by quantitative real-time polymerase chain reaction (qRT-PCR). We used ΔCt method and investigated the differences between tumor tissues and normal colon mucosa by paired t-test. One-way analysis of variance was conducted to analyze the relationship between hsa_circ_001988 expression level and clinic pathological factors of patients. Receiver operating characteristic (ROC) curve was built by SPSS to evaluate the diagnostic values. The expression of hsa_circ_001988 was significantly correlated with differentiation (P<0.05) and perineural invasion (P<0.05). The area under ROC curve of hsa_circ_001988 was 0.788 (P<0.05). Those results indicate that hsa_circ_001988 may become a novel potential biomarker in the diagnosis of colorectal cancer and a potential novel target for the treatment of colorectal cancer.
Keywords: Circular RNA, hsa_circ_001988, colorectal cancer, biomarker, differentiation
Introduction
Colorectal cancer (CRC) is one of most common cancers worldwide, representing almost 10% of the global cancer incidence, leading a high mortality [1]. Although there was several treatment options to colorectal cancer, CRC patient’s survival is still poor because of lacking effective tools for early diagnosis and limited capacity for optimal therapeutic decision making. Thus, it’s valuable to find an ideal biomarker and therapy target to improve the diagnosis rare and treatment.
Circular RNAs are novel RNA molecules which formed mainly by back-splicing with covalently joined 3’- and 5’-ends [2]. Although circular RNA have been reported more than 20 years [3], such molecules mostly were considered as by-products of splicing errors [4,5]. With the development of novel bioinformatics’ approaches and deep sequencing, lots of circular RNAs were found in eukaryotic cells. Recent studies have mainly found three kinds of circular RNAs: exonic circRNA, circular intronic RNA and retained-intron circRNA [5-7]. Several possible functions have been claimed: regulator of translation [7], miRNA sponges [8], binding protein [9] and RNA transport [10]. Some of circular RNAs could act as microRNA sponge to compete endogenous RNA, affecting expression of related gene. The exotic circRNA of CDR1 as (cerebellar degeneration-related protein 1 antisense) have been shown to bind miRNA-7, inducing impaired midbrain development in zebra fish [8]. Furthermore, circular RNAs are related to many diseases [11]. For an example, the abundance of certain circRNA are associated with gastric cancer [12]. However little is known about relationship between the expression level of hsa_circ_001988 and colorectal carcinoma.
We selected hsa_circ_001988 as targeted circular RNA to explore relationship between the expression of this circular RNA and colorectal cancer for following reasons. (1) It’s based on our own next generation sequence data. (2) Hsa_circ_001988 has been predicted by Memczak et al, but they didn’t validate the existence of hsa_circ_001988 [8]. (3) FBXW7 (F-box and WD repeat domain-containing 7), also called hCDC4, is located on chromosomal region 4q3 which is a tumor suppressor [13]. Depletion of FBXW7 is often observed in a variety of tumors including colon cancer [14]. As a result, we firstly validated the existence of hsa_circ_001988 and analyzed association between this circular RNA expression and clinical significances in colorectal cancer patients.
Materials and methods
Patients and clinical information
Following informed patient consent and ethical approval, total of 62 specimens (tumor tissues and adjacent normal mucosa) were obtained from CRC patients during operation and were quickly put into RNA later (Ambion, life technologies, USA). Then we put those specimens into 4°C for 12 h and stored at -80°C until use. Tumors were classified according to the tumor-node-metastasis (TNM) staging system. Histological grade was evaluated following the National Comprehensive Cancer Network clinical practice guideline of oncology (V.1.2012).
Carbohydrate antigen 19-9 (CA19-9) and serum carcinoembryonic antigen (CEA) and were measured using Cobos 8000 (Roche Diagnostics, Basel, Switzerland). Their cutoff values were 35 U/ml and 5 ng/ml respectively.
Total RNA extraction and cDNA synthesis
TRIzol reagent (Ambion, life technologies, USA) was used to extract RNA from the colorectal tissues and normal mucosa, following the manufacturer’s instruction. Then, concentration of RNA was estimated by the NanoDrop ND-8000 (Thermo Fisher Scientific, Wilmington, DE). cDNA was synthesized as following method. First, dilute 1-2 μg RNA into 14 µl RNase-free water in a 0.2 mL PCR tube, then add 1 μL random primers and mix well. Second, put the tubes into a PCR thermocycler and incubate at 70°C for 5 minutes. Third, place the plate on ice immediately and incubate for 2 minutes. Forth, prepare the reaction mix as following: 5 μL 5× Buffer, 1.3 μL dNTP (10 mM), 1 μL M-MLV, 0.6 μL RNase inhibitor, 15μL RNA and random mix, 2.1 μL RNase-free water. Incubate at 42°C for 1 hour, 70°C for 15 minutes and 4°C forever. No template control was conducted at the same time.
Quantitative real-time PCR
Divergent primers were designed with Vector NTI 10. Their sequences were as follows: 5’-CCAGTAGTATTGTGGACCTGCCC-3’ (sense) and 5’-CCCTCTGACCCAGTAACTCCACT-3’ (antisense) for hsa_circ_001988; 5’-TCCTCACAGTTGCCATGTAGACCC-3’ (sense) and 5’-TGCGGGCTCAATTTATAGAAACCGGG-3’ (antisense) for GAPDH. To validate the existence of hsa_circ_001988 in colorectal cancer tissues, the product of quantitative real time PCR was sequenced and the sequencing results were aligned with the theoretical sequence. The expression of hsa_circ_001988 was analyzed by using ΔCt method [15]. All the results are expressed as the mean ± SD of three independent experiments.
Statistical analysis
Statistical data were analyzed using Statistical Program for Social Sciences (SPSS) 17.0 software (SPSS, Chicago, IL, USA). GraphPad Prism 5.0 (GraphPad Software, La Jolla, CA) was used to plot all graphs. One-way analysis of variance was used to analyze the relationship between hsa_circ_001988 expression level and clinic pathological factors in CRC patients. The differences between tumor tissues and normal colon mucosa were analyzed by paired t-test. Receiver operating characteristic (ROC) curve was constructed to evaluate the diagnostic values. P<0.05 was considered statistically significant.
Results
General characteristics of the patients
In order to exclude possible influence of cancer types, we just selected patients diagnosed with adenocarcinoma by HE stain. Figure 1A shows poorly differentiated adenocarcinoma (HE, ×100), Figure 1B shows moderately differentiated adenocarcinoma (HE, ×100), and Figure 1C shows well differentiated adenocarcinoma (HE, ×100). The average of 31 patients with colorectal cancer was 59.10±2.42 years old. The majority of patients with colorectal cancer were man (19, 61.3%). Three patients had perineural invasion (Table 1).
Figure 1.
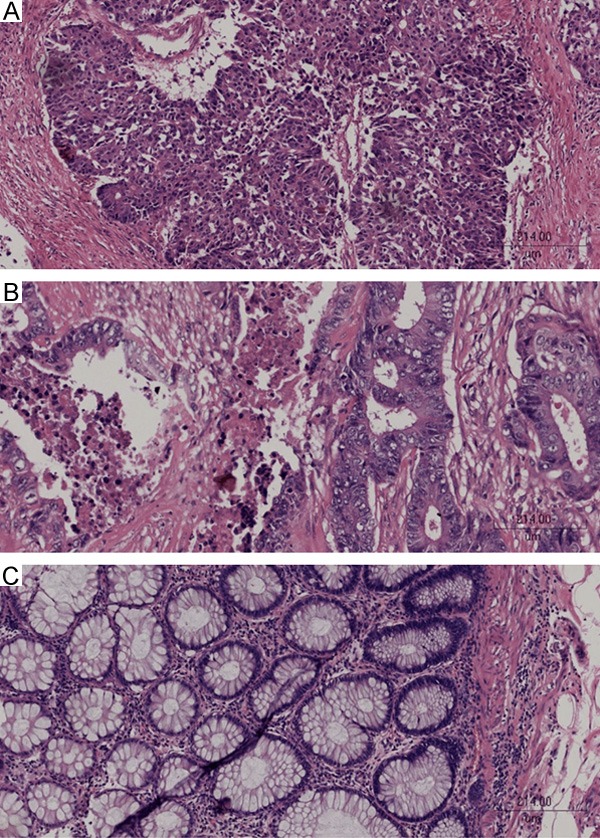
A. Poor differentiation adenocarcinoma, HE, ×100. B. Moderate differentiation adenocarcinoma, HE, ×100. C. Well differentiation adenocarcinoma, HE, ×100.
Table 1.
The relationship of hsa_circ_001988 expression levels (ΔCt) in cancer tissue with clinical features in colorectal cancer patients
| Characteristics | No. Of patients | Mean ± SD | 95% CI | P value |
|---|---|---|---|---|
| Gender | ||||
| Male | 19 | 6.55±0.94 | 6.10-7.00 | P=0.354 |
| Female | 12 | 6.95±1.41 | 6.05-7.84 | |
| Age | ||||
| ≤60 | 15 | 6.47±1.07 | 5.88-7.06 | P=0.280 |
| >60 | 16 | 6.92±1.19 | 6.29-7.55 | |
| Diameter | ||||
| >5 | 8 | 6.31±0.80 | 5.64-6.98 | P=0.261 |
| ≤5 | 23 | 6.84±1.22 | 6.31-7.37 | |
| Differentiation | ||||
| Poor | 7 | 5.59±0.45 | 5.17-6.00 | P=0.008 |
| Moderate | 20 | 7.04±1.16 | 6.49-7.58 | |
| Well | 4 | 6.98±0.56 | 6.10-7.87 | |
| Lymphatic metastasis | ||||
| Nx&N0 | 15 | 6.82±1.29 | 6.10-7.53 | P=0.601 |
| N1&N2 | 16 | 6.60±1.00 | 6.06±7.13 | |
| Invasion | ||||
| Tis&T0-T3 | 24 | 6.67±1.12 | 6.19-7.14 | P=0.748 |
| T4 | 7 | 6.82±1.28 | 5.64-8.01 | |
| TNM stage | ||||
| 0&I&II | 15 | 6.82±1.28 | 6.10-7.53 | P=0.601 |
| III&IV | 16 | 6.60±1.00 | 6.06-7.13 | |
| PNI | ||||
| Negative | 28 | 6.84±1.11 | 6.41-7.26 | P=0.037 |
| Positive | 3 | 5.42±0.38 | 4.46-6.37 | |
| CEA | ||||
| Positive | 14 | 6.99±1.37 | 6.20-7.78 | P=0.202 |
| Negative | 17 | 6.46±0.87 | 6.01-6.91 | |
| CA19-9 | ||||
| Positive | 6 | 6.97±1.64 | 5.24-8.70 | P=0.527 |
| Negative | 25 | 6.63±1.01 | 6.21-7.05 |
Existence of hsa_circ_001988 in colorectal cancer tissues
Divergent primers were used to amplify circular RNA. The product of quantitative real time PCR was sequenced and the sequence of qRT-PCR’s product could cross cleavage site. Those results validated the existence of hsa_circ_001988 in the tissues.
Hsa_circ_001988 expression and clinicopathologic factors in colorectal cancer patients
Compared with normal samples, the expression of hsa_circ_001988 was significantly down regulated (Figure 2). To further explore the relationship between hsa_circ_001988 and clinical features, we analyzed following clinic pathological factors with colorectal cancer (Table 1). The expression level of hsa_circ_001988 was related to differentiation (P<0.05) and perineural invasion (P<0.05). Many other clinic pathological features were analyzed, but those features (gender, age, diameter, lymphatic metastasis, TNM stage et al) were not associated with hsa_circ_001988 according to statistical analysis.
Figure 2.
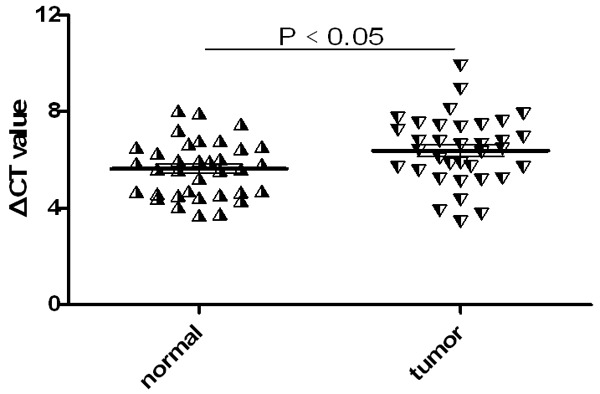
The expression levels of hsa_circ_001988 in colorectal cancer patients. The expression levels of hsa_circ_001988 in 31 matched carcinoma and normal samples. Higher ΔCt value indicates lower expression.
ROC curve of hsa_circ_001988
To estimate whether hsa_circ_001988 could be used as an indicator for diagnosis of colorectal cancer, a ROC curve was built. A total of 31 patients’ normal colon mucosa was used as a control to produce this ROC curve (Figure 3). The sensitivity and specificity was 0.68 and 0.73, respectively. The cutoff value was 6.04. The area under the curve was 0.788 (95% CI=0.68-0.90, P<0.001). The Youden index was 0.419. Therefore, hsa_circ_001988 could be used as indictor of colorectal cancer.
Figure 3.
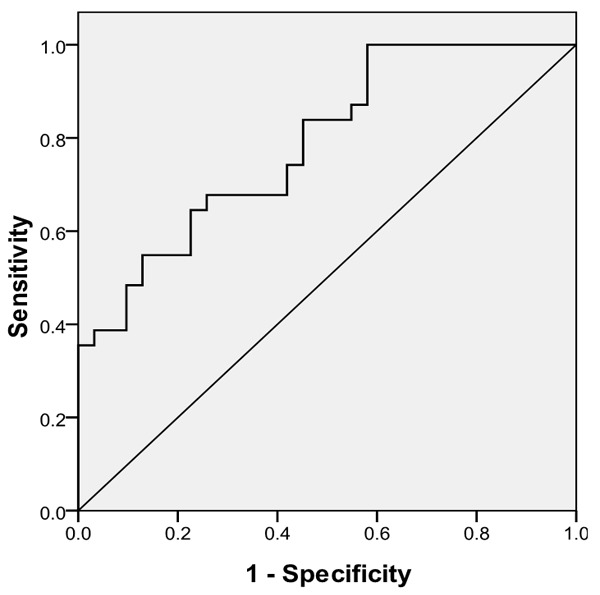
The ROC curve was constructed by SPSS. Area under curve was 0.788. The sensitivity and specificity was 0.68 and 0.73, respectively. P<0.05.
Discussion
Circular RNAs are newly found endogenous noncoding RNAs, which had been considered as the results of transcriptional noise [4]. With the development of bioinformatics’ methods and sequencing technology, lots of circular RNAs were predicted. Many researchers show great interest in those noncoding RNAs to explore their function. For an example, CDR1 as/ciRS-7 and Sry, which have been studied a lot, act as microRNA sponges [8,16], while other studies have speculated that circRNAs may have other functions, which may be related to protein and/or RNA transport [10].
Many further studies about relationship between circular RNA and disease were conducted. Such as, Lukiw et al reported CDR1 as associated with Alzheimer’s disease [17]. Cyclization of an INK4/ARF-Associated non-coding RNA correlates with atherosclerosis risk [18]. And the study conducted by Ghosal et al. found 105 diseases were connected with circular RNAs from 174 diseases including several carcinomas and neurodegenerative diseases [11]. However it was little known about the relationship between hsa_circ_001988 and colorectal cancer. To our knowledge, this is the first report of the existence of hsa_circ_001988. Our findings showed that the expression of hsa_circ_001988 was decreased in tumor tissues compared with normal mucosa and the average hsa_circ_001988 expression of PNI-positive tumors is lower than PNI-negative tumors (Table 1). Furthermore, the expression level of hsa_circ_001988 was significantly associated with perineural invasion (P<0.05) and differentiation (P<0.05).
It has been identified perineural invasion is a predictor of outcome in colorectal cancer and perineural invasion was negatively associated with survival time and local recurrence in colorectal cancer patients [19,20]. Besides, the degree of tumor cell differentiation is associated with prognosis. Thus, hsa_circ_001988 may play a profound role in differentiation and perineual invasion. It is a potential target to regulate cytological behaviors. Furthermore, hsa_circ_001988 may be used as new target to therapy colorectal cancer and predict colorectal cancer cell differentiation and perneural invasion.
However, our study just validated the existence of hsa_circ_001988 and analyzed the relationship of between this circular RNA and clinical features in 31 patients. More samples are needed in the future work. And it’s necessary to explore the further function of hsa_circ_001988.
In conclusion, hsa_circ_001988 could be detected in human colorectal cancer tissues and normal mucosa. The expression of hsa_circ_001988 was down regulated in colorectal cancers. Furthermore, its expression was associated with differentiation and perinerual invasion. Those results suggest that hsa_circ_001988 may be a new target for treatment of colorectal cancer and a potential biomarker of colorectal cancer.
Acknowledgements
This work was supported by 863 Program of China (No. 2014AA020802).
Disclosure of conflict of interest
None.
References
- 1.Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D, Bray F. Reprint of: Cancer incidence and mortality patterns in Europe: Estimates for 40 countries in 2012. Eur J Cancer. 2015;51:1201–1202. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 2.Hentze MW, Preiss T. Circular RNAs: splicing’s enigma variations. EMBO J. 2013;32:923–925. doi: 10.1038/emboj.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B. Scrambled exons. Cell. 1991;64:607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- 4.Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 5.Danan M, Schwartz S, Edelheit S, Sorek R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012;40:3131–3142. doi: 10.1093/nar/gkr1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 8.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 9.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 11.Hancock JM. Circles within circles: commentary on Ghosal et al. (2013) “Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits”. Front Genet. 2014;5:459. doi: 10.3389/fgene.2014.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li P, Chen S, Chen H, Mo X, Li T, Shao Y, Xiao B, Guo J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136. doi: 10.1016/j.cca.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 13.Crusio KM, King B, Reavie LB, Aifantis I. The ubiquitous nature of cancer: the role of the SCF (Fbw7) complex in development and transformation. Oncogene. 2010;29:4865–4873. doi: 10.1038/onc.2010.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwatsuki M, Mimori K, Ishii H, Yokobori T, Takatsuno Y, Sato T, Toh H, Onoyama I, Nakayama KI, Baba H, Mori M. Loss of FBXW7, a cell cycle regulating gene, in colorectal cancer: clinical significance. Int J Cancer. 2010;126:1828–1837. doi: 10.1002/ijc.24879. [DOI] [PubMed] [Google Scholar]
- 15.Schmittgen TD, Livak KJ. Analyzing realtime PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 16.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 17.Lukiw WJ. Circular RNA (circRNA) in Alzheimer’s disease (AD) Front Genet. 2013;4:307. doi: 10.3389/fgene.2013.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang HY, Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of Linear and Novel Circular Forms of an INK4/ARFAssociated Non-Coding RNA Correlates with Atherosclerosis Risk. PLoS Genetics. 2010;6:e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liebig C, Ayala G, Wilks J, Verstovsek G, Liu H, Agarwal N, Berger DH, Albo D. Perineural invasion is an independent predictor of outcome in colorectal cancer. J. Clin. Oncol. 2009;27:5131–5137. doi: 10.1200/JCO.2009.22.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng J, Sheng W, Huang D, Venook AP, Xu Y, Guan Z, Cai S. Perineural invasion in pT3N0 rectal cancer: the incidence and its prognostic effect. Cancer. 2011;117:1415–1421. doi: 10.1002/cncr.25620. [DOI] [PubMed] [Google Scholar]


