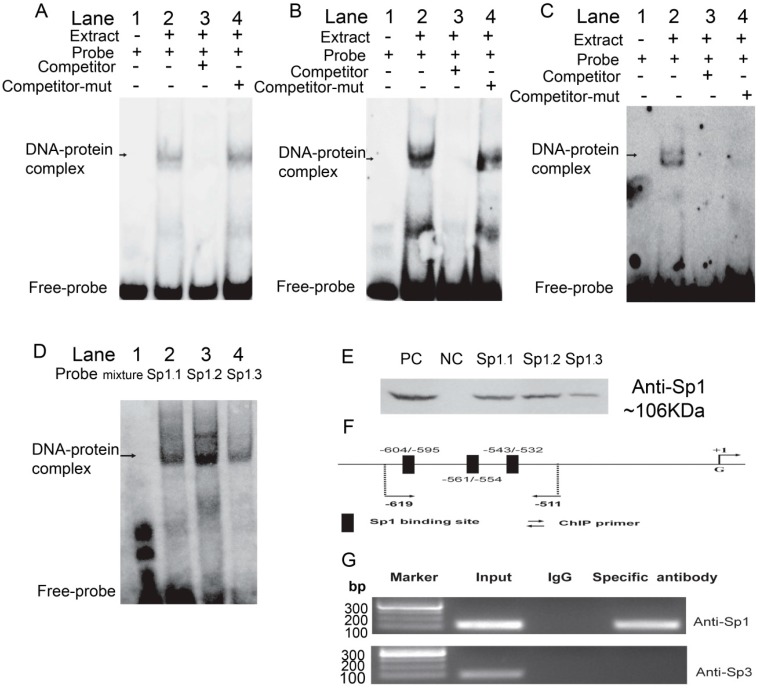
Figure 3.
Binding of Sp1 with the ROCK1-P5 fragment was analyzed in vitro and in vivo. The first (A), the second (B) and the third (C) biotin-labeled probes were incubated with the NE of PK cells. Lane 1 was the negative control without NE; the reagents were incubated in the absence competitor probes in Lane 2 or in presence of 50× excess competitor (Lane 3)/competitor-mutant (Lane 4) probes, respectively; (D) The three probes were incubated with PK NE, respectively; (E) Proteins of PK extracted from DNA-pull down materials were detected by Western blot. The total non-denaturing proteins/Streptavidin MagneSphere® Paramagnetic Particles were taken as positive/negative control (PC/NC). The three potential Sp1 binding sites were named as Sp1.1, Sp1.2, and Sp1.3 in (D,E). The competitor/competitor-mutant probes were 50-fold excess and arrows indicated the specific DNA-protein complex bands; (F) Schematic diagram of the Sp1 binding sites in the porcine ROCK1-P5 fragment; (G) ChIP assay of Sp1 binding to porcine ROCK1-P5 fragment in PK cells. The in vivo interaction of Sp1 and Sp3 with porcine ROCK1 promoter was determined by ChIP assay, in which Normal mouse IgG was used as negative control. DNA isolated from immunoprecipitated materials was used for PCR amplification, whereas total chromatin was used as input (positive control). The antibodies used in ChIP assay were listed in the right of the figure and the corresponding amplification product obtained here was 107 bp.