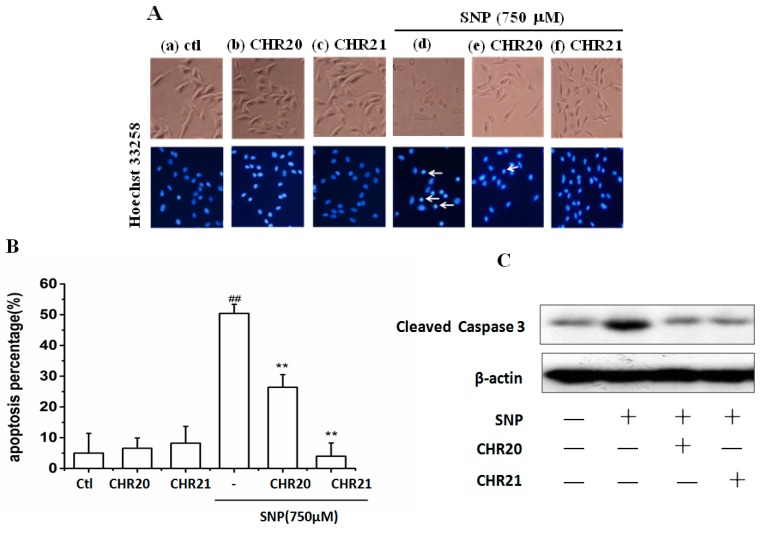
Figure 5.
CHR20 and CHR21 protected RGC-5 cells from apoptosis induced by SNP. RGC-5 cells were pretreated with/without CHR20/CHR21 for 2 h, respectively, after treatment of 750 μM SNP for 24 h and the cells were stained with Hoechst 33258 as described in Materials and Methods. (A) Morphological changes shown by fluorescence microscope (200×) image analysis. (a) control group; (b) 30 μM CHR20 group; (c) 30 μM CHR21 group; (d) 750 μM SNP group; (e) 750 μM SNP + 30 μM CHR20 group; (f) 750 μM SNP + 30 μM CHR21 group. The arrow indicates nuclear fragmentation or chromatin condensation; (B) Histogram showing the apoptosis rate in RGC-5 cells. The number of apoptic cells was about 1.5 × 104 cells/well in 25 visual fields in 48-well culture plate, counted by high content screening system (ArrayScanVTI, Thermo Fisher Scientific, Waltham, MA, USA). The percentage of apoptotic cells was calculated as ratio of apoptotic cells and the total number of cells counted. Data are given as mean ± SD for three individual experiments. ## p < 0.05 vs. control group. ** p < 0.05 vs. SNP group; (C) Effects of CHR20/CHR21 on cleaved caspase-3. The expression of cleaved caspase-3 was determined by western blotting as described in Materials and Methods.