SUMMARY
The ability to sense energy status is crucial in the regulation of metabolism via the mechanistic Target of Rapamycin Complex 1 (mTORC1). The assembly of the TTT-Pontin/Reptin complex is responsive to changes in energy status. In energy sufficient conditions, the TTT-Pontin/Reptin complex promotes mTORC1 dimerization and mTORC1-Rag interaction, which are critical for mTORC1 activation. We show that WAC is a regulator of energy-mediated mTORC1 activity. In a Drosophila screen designed to isolate mutations that cause neuronal dysfunction, we identified wacky, the homolog of WAC. Loss of Wacky leads to neurodegeneration, defective mTOR activity and increased autophagy. Wacky and WAC have conserved physical interactions with mTOR and its regulators, including Pontin and Reptin which bind to the TTT complex to regulate energy-dependent activation of mTORC1. WAC promotes the interaction between TTT and Pontin/Reptin in an energy-dependent manner, thereby promoting mTORC1 activity by facilitating mTORC1 dimerization and mTORC1-Rag interaction.
Keywords: Wacky, Drosophila, autophagy, energy sensing, neurodegeneration
INTRODUCTION
The ability to modulate catabolic and anabolic processes in response to fluctuations in nutrient and energy availability is essential for an organism’s survival. In eukaryotes, the serine/threonine kinase, mechanistic Target of Rapamycin (mTOR), is the central player in the regulation of growth and metabolism in response to environmental cues. mTOR is an integral part of two complexes, mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2), with two complex-specific adaptors, Raptor and Rictor, respectively. mTORC1 responds to intracellular and extracellular cues, including energy status, growth factors, and amino acids, and positively regulates protein and lipid synthesis and energy metabolism while negatively regulating autophagy. mTORC2 responds to growth factors and regulates cell survival and metabolism as well as cytoskeletal organization (Kim and Guan, 2015; Laplante and Sabatini, 2012). The complex interplay between environmental cues and metabolism hinges on mTORC1, but the mechanisms by which signals, especially nutrient availability, are sensed and integrated by mTORC1 are still being elucidated (Figure S1A).
mTORC1 has been shown to be activated by the Rag and Rheb GTPases in essential but distinct ways. In response to amino acid availability, mTORC1 is recruited by Rag GTPases to the lysosomal surface (Kim et al., 2008, 2013a; Laplante and Sabatini, 2012; Sancak et al., 2010). Permissive growth factor signaling releases the lysosomally-localized Rheb GTPase from inhibition by the GTPase activating protein (GAP) complex, Tuberous Sclerosis Complex (TSC)1/2, thereby enabling Rheb to stimulate mTOR kinase activity (Inoki et al., 2003a; Tee et al., 2003).
mTORC1 has also been shown to be negatively regulated by decrease in energy levels through the activation of AMP-activated Protein Kinase (AMPK). AMPK, which responds to increased AMP/ATP ratio, phosphorylates TSC2 and Raptor, which in turn activates TSC2 function and decreases Raptor activity, leading to mTORC1 inhibition (Gwinn et al., 2008; Inoki et al., 2003b).
Recently, mTORC1 was found to positively respond to increased energy availability in a TSC1/2-, AMPK-, and Rag-independent manner through regulation by the TTT-Pontin/Reptin complex (Kim et al., 2013b). The AAA+ (ATPase associated with diverse cellular activities) ATPases Pontin/Reptin (RUVBL1/RUVBL2) complex functions as a chaperone that modulates proper folding of large proteins as well as assembly of protein complexes (Huen et al., 2010). This complex is known to bind the TTT complex composed of TEL2, TTI1 and TTI2. These complexes form the larger TTT-Pontin/Reptin complex that controls the energy-dependent assembly of functional mTORC1, its dimerization, and its association with Rag for lysosomal localization (Kim et al., 2013b). Energy depletion, through deprivation of glucose and glutamine, the primary carbon sources for the ATP-producing tricarboxylic acid cycle, leads to disassembly and repression of the TTT-Pontin/Reptin complex. This in turn leads to disruption of mTORC1 dimerization and of mTORC1-Rag interaction. However, little is known about how the energy-dependent assembly of the TTT-Pontin/Reptin complex is regulated.
In this study we identified the Drosophila homolog of WAC, which we have named wacky, as a regulator of the mTOR pathway. wacky mutants exhibit phenotypes that are characteristic of impaired mTOR signaling, including developmental arrest, decreased cellular growth and cell size, and increased autophagy. Wacky is associated with dTOR, Pontin, and Reptin. In HEK 293T cells, depletion of WAC similarly compromises mTOR signaling and increases autophagy. WAC also physically interacts with mTORC1 and subunits of the TTT and Pontin/Reptin complexes in an energy-dependent manner. WAC promotes the energy-dependent interaction between the individual TTT and Pontin/Reptin complexes to form the larger, functional TTT-Pontin/Reptin complex facilitating mTORC1 dimerization and mTORC1-Rag interaction. We propose that WAC promotes mTOR activity by acting as an adaptor for the proper assembly of the TTT-Pontin/Reptin complex.
RESULTS
XE14 alleles affect neuronal function, survival, and basal autophagy
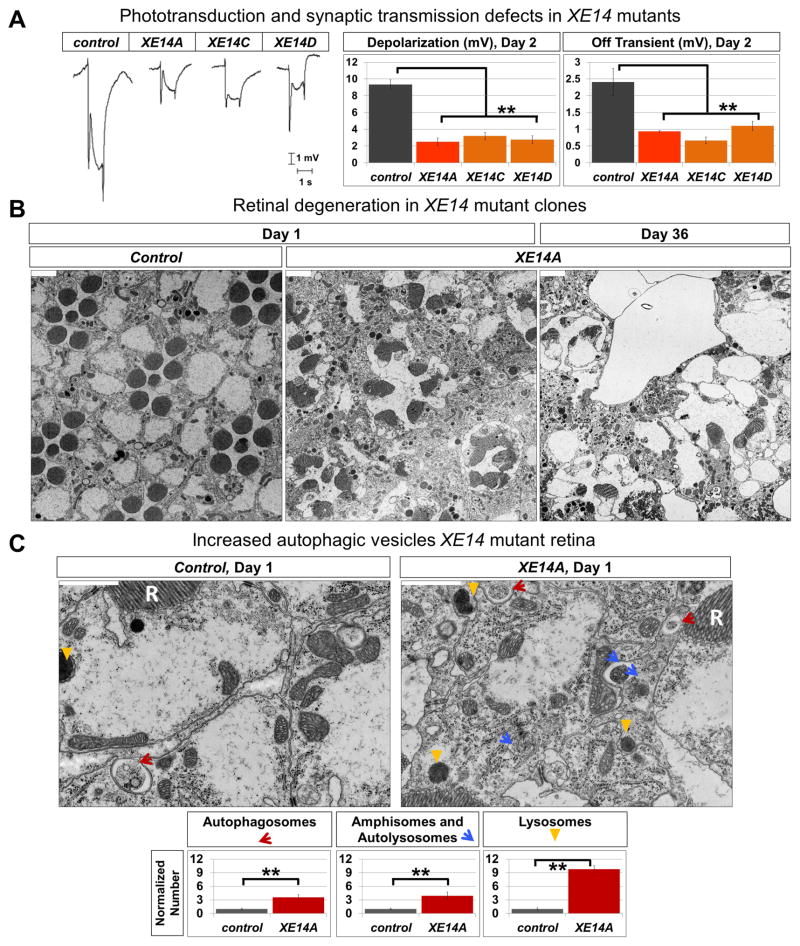
To identify essential genes involved in the function and maintenance of the nervous system in Drosophila, we performed an unbiased mutagenesis screen on the X chromosome using ethyl methane-sulfonate (Haelterman et al., 2014; Yamamoto et al., 2014). We generated mosaic eyes that contain homozygous mutant cells using the FLP-FRT system (Xu and Rubin, 1993) and recorded electroretinograms (ERGs) from mutant clones (Jaiswal et al., 2015; Sandoval et al., 2014; Tian et al., 2015; Wang et al., 2014; Xiong et al., 2012; Zhang et al., 2013). The ERG response consists of a depolarization, which is a measure of the strength of phototransduction, and the on and off transients, which gauge synaptic transmission. From this screen, we identified five alleles of a complementation group, XE14, that cause similar defects in phototransduction and synaptic transmission in mutant clones (Figure 1A).
Figure 1. XE14 alleles affect neuronal function, survival, and basal autophagy.
(A) ERG traces from 2-day old ey-FLP clones of control (y w FRT19A) (n=6) and XE14 alleles (n=6), with quantification for ERG amplitude and Off transient. Error bars are SEM and ** p<0.02.
(B) Transmission electron microscope (TEM) images of the retina of ey-FLP clones of control (y w FRT19A) and XE14A raised in 12-hour light/12-hour dark cycle, at 1 day old and 36 days old. Scale bars are 2 μm.
(C) Higher magnification TEM images of the retina of ey-FLP clones of control (y w FRT19A) (n=3) and XE14A (n=3), raised in 12-hour light/12-hour dark cycle, at 1 day old, with quantification of autophagic vesicles. R stands for rhabdomere. Scale bars are 1 μm, error bars are SEM, and ** is p<0.02.
See also Figure S1.
To determine if the defective ERG responses exhibited by XE14 alleles are associated with structural defects, we performed transmission electron microscopy (TEM) of adult retinae and laminae, which revealed an age-dependent degeneration of internal eye structures (Figures 1B and S1B). At day 1, XE14 mutant retinae show obvious disruptions of ommatidial units, with abnormally fused or split rhabdomeres and loss of photoreceptor cells. By day 36, these defects are more severe and numerous photoreceptors are lost, leaving large vacuoles, suggesting cell death. Closer examination of 1-day old fly retinae reveals marked increase in autophagosomes, amphisomes, autolysosomes, and lysosomes (classified according to Eskelinen, 2008; Klionsky et al., 2012; Lullmann-Rauch, 2005) when compared to control (Figure 1C). Similar structural abnormalities are observed in 1-day old photoreceptor terminals in the laminae of XE14 mutants, showing increase in autophagic vesicles as well as marked disorganization and expansion of cartridge structures (Figure S1C).
XE14 encodes the Drosophila WAC homolog, Wacky, a broadly-expressed protein localized to lysosomes and nuclei
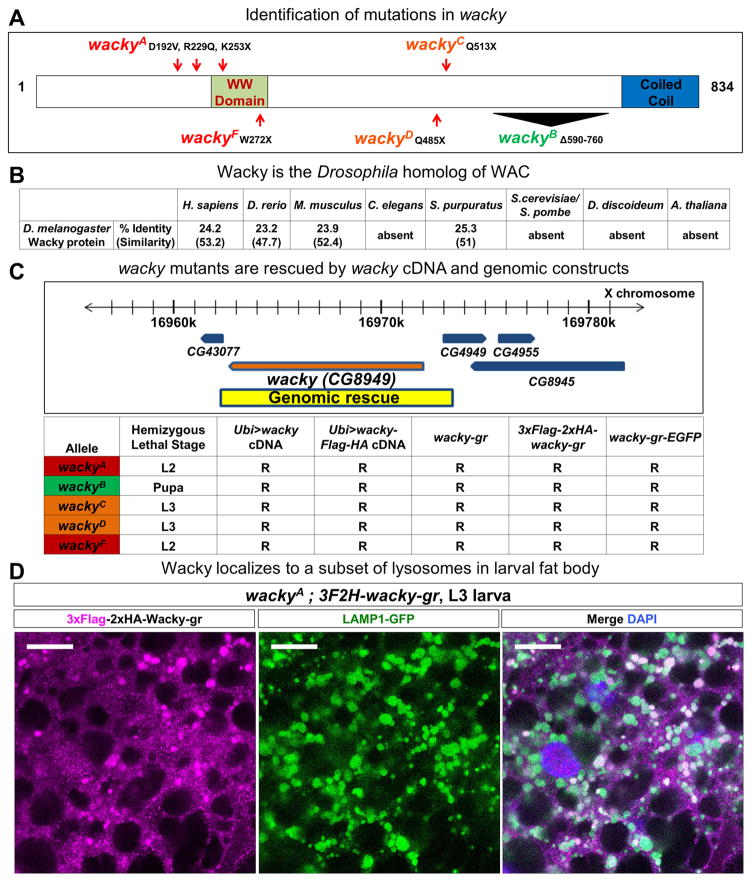
We mapped the gene that corresponds to XE14 to CG8949 encoding a putative 834-amino acid protein with sequence homology to human WAC (24.2% identity and 53.2% similarity) (Figure 2A). WAC contains a WW domain and a coiled coil region, two protein-binding domains for which it was named. Interestingly, WAC is conserved in human, mouse, and zebrafish, but absent in lower eukaryotes such as C. elegans and yeasts (Figure 2B). Because the gene symbol, wac, corresponds to wee Augmin, we renamed CG8949 wacky.
Figure 2. XE14 encodes the Drosophila WAC homolog, wacky, a broadly-expressed protein with nuclear and lysosomal localization.
(A) Schematic representation of Wacky molecular lesions identified in XE14 alleles.
(B) Conservation of Wacky in humans and several model organisms.
(C) wacky cDNA constructs and genomic (gr) constructs, spanning the genomic region indicated, rescue wacky lethality. R, rescued.
(D) Immunofluorescence staining with FLAG antibody of wackyA rescued with 3xFLAG-2xHA-tagged genomic wacky transgene in the fat body of wandering L3 larvae. Scale bars are 20 μm.
See also Figure S2.
To ensure that the phenotypes observed for wacky mutants are due to mutations in the wacky gene, we generated both untagged and tagged wacky genomic (gr) and cDNA rescue constructs (Figure 2C). Introduction of the genomic constructs or ubiquitous expression of wacky cDNA in wacky mutants rescued lethality, ERG defects, and morphological alterations associated with wacky alleles (Figures 2C, S2A, and S2B). By using a 3xFlag-2xHA amino terminal-tagged genomic wacky construct, we observed that Wacky is broadly expressed in adult and larval tissues (Figures S2C and S2D) and is present in the nucleus as well as a subset of lysosomes in the fat body cells of wandering third instar larvae (Figure 2D).
Loss of Wacky promotes basal autophagy in Drosophila
In mammalian cells, WAC has been found to be a positive regulator of autophagy, as siRNA-mediated knockdown of WAC in HEK 293T cells blocks starvation-induced autophagosome formation, LC3 lipidation, and p62 turnover (McKnight et al., 2012), a finding that we have also confirmed (data not shown). In Drosophila, the larval fat body is a well-established system to study autophagy (Chang and Neufeld, 2010; Neufeld and Baehrecke, 2008; Zirin and Perrimon, 2010). In second and early third instar larvae, fat body cells display minimal autophagy activity under well-fed conditions but show robust autophagy induction upon starvation, with prominent accumulation of autophagosomes and lysosomes. In agreement with findings that WAC positively regulates starvation-induced autophagy, clones of wacky mutant cells from starved animals show minimal autophagy induction compared to surrounding normal cells, a phenotype that can be fully rescued by the wacky genomic rescue construct (Figure S3A).
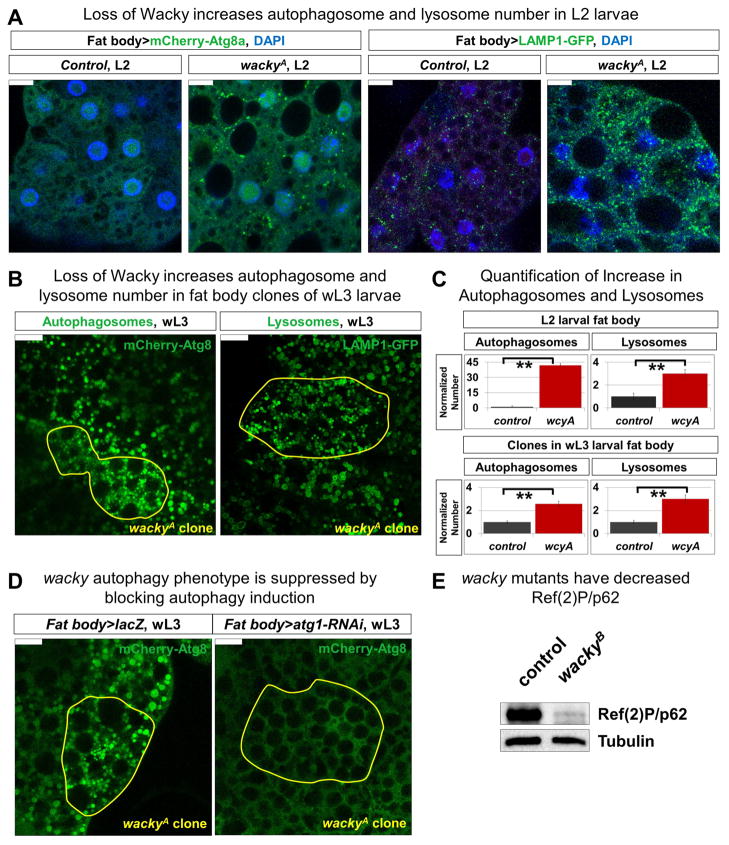
However, the increase of autophagic vesicles in wacky mutant retina suggests that Wacky is also a negative regulator of autophagy. Hence, we explored its role in autophagy regulation under basal conditions. In addition to starvation-induced autophagy, fat body cells also undergo developmentally-programmed autophagy, showing minimal levels of autophagy in well-fed second and early third instar larvae but a strong increase of autophagic activity in late third instar larvae in response to bursts of the steroid hormone, ecdysone, prior to metamorphosis (Rusten et al., 2004; Scott et al., 2004). Similar to what we observed in clones of mutant adult retinae, fed wackyA mutants show increase in the number of autophagosomes and lysosomes in fat body cells of second instar larvae, as revealed by mCherry-Atg8a (Nezis et al., 2009) and LAMP1-GFP (Pulipparacharuvil et al., 2005), markers for autophagosomes and lysosomes, respectively (Figures 3A and 3C). Furthermore, co-expression of these markers in wacky mutant fat body shows predominant co-localization of mCherry-Atg8a and LAMP1-GFP in these cells, indicating that loss of wacky does not affect autophagosome-lysosome fusion (Figure S3B).
Figure 3. Wacky is a negative regulator of basal autophagy in Drosophila.
(A) Control (y w FRT19A) and wackyA second instar larval fat body expressing UAS-mCherry-Atg8a (to mark autophagosomes) and UAS-LAMP1-GFP (to mark lysosomes) with the fat body-specific Cg-GAL4 driver. Scale bars are 10 μm.
(B) wackyA clones, marked yellow, in wandering third instar larval fat body expressing UAS-mCherry Atg8a and LAMP1-GFP with Cg-GAL4 driver. Scale bars are 20 μm.
(C) Quantification of autophagosome and lysosome numbers from (A) (n=4), and (B) (n=5). Error bars are SEM and ** is p<0.02.
(D) Cg-GAL4-driven expression of UAS-lacZ as control or UAS-Atg1-RNAi to suppress autophagy induction in third instar larval fat body with wackyA clones marked yellow. Scale bars are 20 μm.
(E) Western blot for Ref(2)P/p62 with fat body protein lysates from third instar larval wackyB ; wacky-gr (control) and wackyB.
See also Figure S3.
To test whether increased autophagy in wacky mutants is cell autonomous, we generated wackyA mutant clones in fed larval fat body and looked at fat body-expressed mCherry-Atg8a and LAMP1-GFP. At the early third instar larval stage, when autophagic activity is minimal in surrounding normal cells, mutant clones show marked increase of both autophagosomes and lysosomes (data not shown). Similarly, at the wandering third instar larval stage, when autophagic activity becomes readily observable, mutant clones exhibit increased number as well as decreased size of autophagosomes and lysosomes, supporting an autonomous role of wacky in autophagy regulation (Figures 3B and 3C). Importantly, the increased number of autophagic structures in wackyA fat body mutant clones is abolished upon RNAi-mediated knock down of Atg1, a kinase required for autophagy initiation, directed by the fat body-Gal4 driver, Cg-Gal4 (Figure 3D). These data show that the increase in the number of autophagic structures in wackyA mutants is due to increase in autophagy. Consistent with these data, we observe marked reduction of the level of Ref(2)P, the Drosophila homolog of the autophagy substrate p62, in the fat body of wackyB mutant larvae (Figure 3E). Together, these findings support a role of WAC in differentially regulating autophagy under different settings, potentially through distinct mechanisms (see Discussion), and suggest that Wacky acts as a negative regulator of basal autophagy in Drosophila.
Wacky functionally interacts with dTOR in Drosophila
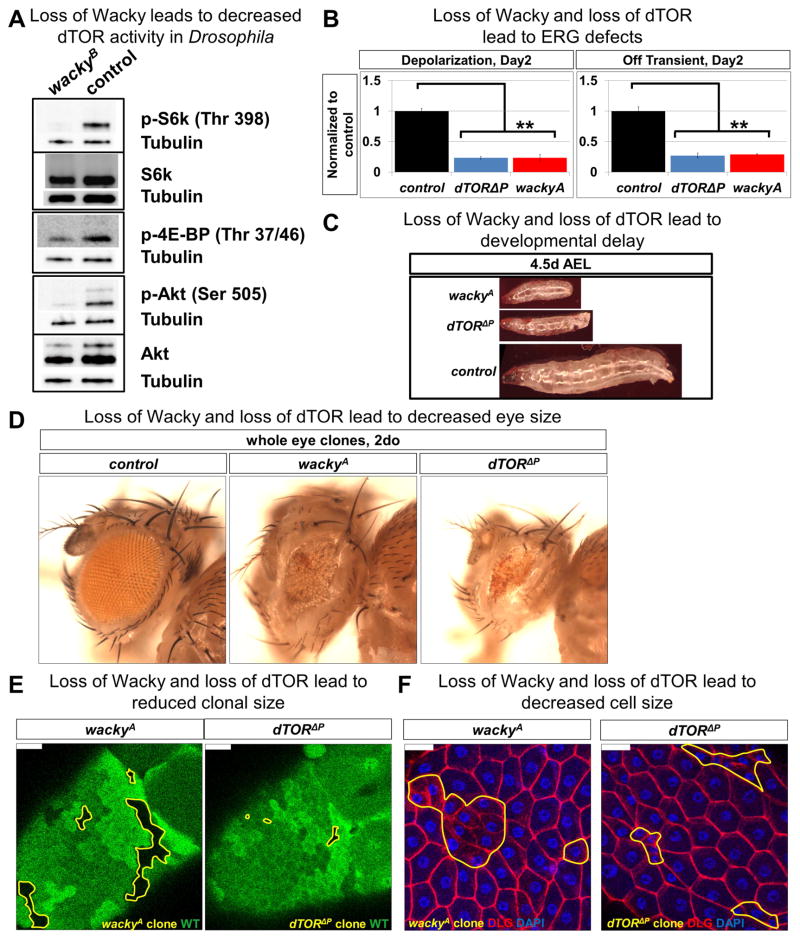
To determine how Wacky is regulating basal autophagy, we performed proteomic analysis using wacky mutant animals rescued by either the GFP-tagged (experimental group) or untagged (negative control) wacky genomic constructs (Figures 2C, S2A, S2B, S3A, and S3C). We affinity-purified whole larval extracts using GFP nanobody, followed by protein identification by mass spectrometry (AP-MS) (Neumüller et al., 2012) . The top hit among isolated Wacky interactors is Bre1, the fly homolog of established WAC-binding partners, RNF20 and RNF40 (Zhang and Yu, 2011) (Table S1). Intriguingly, among the other specific hits is the Drosophila mTOR homolog, dTOR, as well as homologs of known mTOR regulators Pontin, Reptin, and Vha100-2 (Table S1). Vha100-2 is a component of the vacuolar ATPase (v-ATPase), a proton pump required for the lysosomal localization and activation of mTORC1 (Zoncu et al., 2011), while Pontin and Reptin form a complex that interacts with mTOR (Izumi et al., 2010) and regulates the energy-dependent functional assembly of mTORC1 (Kim et al., 2013b) (Figure S1A). Notably, in Drosophila, dTOR has been shown to interact with the fly homologs of Pontin and Reptin together with the TTT components, further supporting their conserved physical interaction (Glatter et al., 2011). Unfortunately, a dearth of reagents to detect the fly homologs of these mTOR pathway components precluded us from further validating their physical interaction with endogenous Wacky in Drosophila. However, to explore the functional significance of these potential protein interactions, we examined the phosphorylation status of RPS6-p70-protein kinase (S6k) and Eukaryotic translation initiation factor 4E binding protein (4E-BP), as well as Akt, the direct downstream targets of dTORC1 and dTORC2, respectively. Remarkably, in wacky mutants, the phosphorylation levels of these dTOR targets are dramatically diminished, while the levels of total S6K and AKT show only slight reductions (Figures 4A and S4A), suggesting a general reduction of dTOR activity. Consistent with this, phenotypic analyses reveal similarities between wacky and dTOR mutants. Similar to what is observed in wackyA, dTOR P mutants show defective phototransduction and synaptic transmission (Figure 4B). Additionally, wackyA and dTOR P mutant animals exhibit slow growth and developmental delay (Figure 4C), and wackyA and dTOR P whole eye clones are significantly smaller than control (Figure 4D). Furthermore, clones of mutant cells in egg chamber follicle cells show reduced clone and cell size (Figures 4E, 4F, S4D, and S4E), resembling, but milder than, what had been previously observed in dTOR mutants (LaFever et al., 2010; Oldham et al., 2000; Zhang et al., 2000). In summary, our data indicate that Wacky is an important player in dTOR-regulated processes and is part of a complex of proteins that interact with dTOR.
Figure 4. Wacky functionally interacts with mTOR in Drosophila.
(A) Western blot for dTOR-dependent phosphorylation of S6k, 4E-BP, and Akt with fat body protein lysates from third instar larval wackyB ; wacky-gr (control) and wackyB.
(B) ERG traces from 2-day old whole-eye clones of control (FRT40A) (n=5), dTOR P (n=5), and wackyA (n=4), with quantification for ERG amplitude and Off transient. Error bars are SEM and ** p<0.02.
(C) Images of FRT40A (control), wackyAFRT19A, and dTOR P FRT40A larvae at 4.5 days after egg laying (AEL).
(D) Images of whole eye clones of 2-day old control (FRT40A), wackyA, and dTOR P.
(E) wackyA and dTOR P mutant follicle cell clones, marked yellow, in stage 10 egg chambers. Scale bars are 20 μm.
(F) wackyA and dTOR P mutant follicle cells, marked yellow in stage 14 egg chambers. Scale bars are 20 μm.
See also Figure S4.
To test the interaction between Wacky and the dTOR pathway genetically, we overexpressed dTOR and several of its upstream activators in the fat body of wackyA mutant clones. Curiously, while ectopic expression of dTOR from the UAS-dTOR transgene, maintained at 18°C to achieve low level of dTOR expression and avoid toxicity associated with dTOR overexpression (Hennig and Neufeld, 2002), improved the synaptic transmission defects and suppressed autophagosome increase in wackyA mutant clones in retina and fat body (Figures S5B and S5C), none of the tested upstream dTOR activators, including Rheb (Patel and Tamanoi, 2006), a constitutively-active form of Rag (Rag-CA) (Kim et al., 2008), and the dTORC1-specific adaptor Raptor (Wang et al., 2012), showed any effect. These data suggest that a mechanism for dTOR activation, insensitive to Rheb and Rag, is compromised in wacky mutants.
Human WAC regulates mTOR activity and autophagy
The isolated Wacky interactors, Pontin and Reptin, form a complex that controls energy-dependent mTORC1 activation through a Rheb- and Rag-independent mechanism (Kim et al., 2013b). Pontin and Reptin represent an intriguing target for Wacky-mediated regulation of dTOR. Given the scarcity of reagents for this pathway in Drosophila, we turned to human cell culture. When tested in multiple cell lines, RNAi-mediated knockdown of WAC (transient siWAC and stable WAC(−)) results in significantly higher levels of basal autophagy. For HEK293T, HeLa, and neuronal SH-SY5Y cells cultured in rich medium, inhibition of lysosomal degradation by lysosomal protease inhibitors results in higher levels of LC3-II accumulation, indicating increased LC3-II synthesis in these WAC knockdown cells (Figures 5A and S5A). Similarly, in HEK 293T cells, WAC depletion leads to increased fragmentation of the GST-BHMT reporter (Figures 5B and S5B), an end-point cargo-based assay for autophagy (Dennis and Mercer, 2009; Furuya et al., 2001; Ueno et al., 1999). This increase in autophagy is dependent on autophagy induction, as inclusion of 3-methyladenine (3-MA), an inhibitor of class III phosphatidylinositol 3-kinase (PI3K), VPS34, or knockdown of ATG7 (Feng et al., 2014), blocks the WAC knockdown-induced fragmentation of GST-BHMT (Figures 5B and S5B). Hence, the data in human cells are consistent with the data in Drosophila. Indeed, ubiquitous expression of the human WAC cDNA rescues lethality of wackyA (data not shown), confirming the functional conservation of WAC proteins between fly and human.
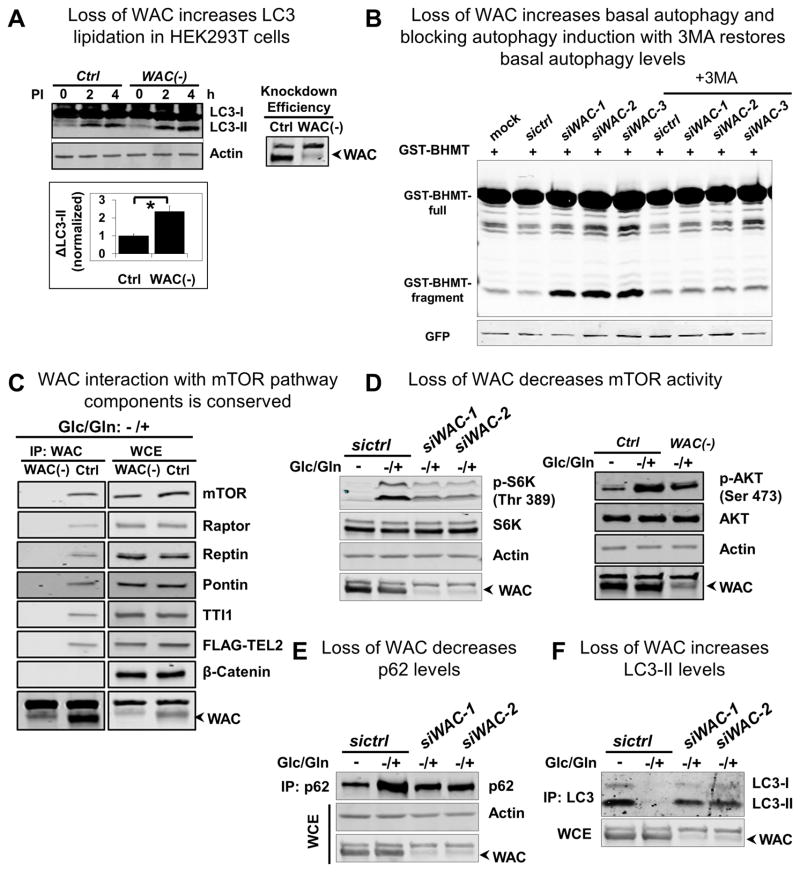
Figure 5. Human WAC regulates mTOR activity and autophagy and interacts with mTORC1 and the TTT-Pontin/Reptin complex.
(A) Basal autophagy is assayed in HEK 293T cells by examining LC3-II levels in the absence or presence of lysosomal protease inhibitors (PI). WAC knock down increases LC3-II levels, indicating an increase in basal autophagy. LC3-II is calculated as the difference between LC3-II levels at 4 hours and 2 hours after PI treatment. Error bars are SEM, n=3, and * p<0.05.
(B) Basal autophagy is assayed in HEK 293T cells by monitoring the autophagy-dependent fragmentation of the GST-BHMT reporter. WAC knock down increases the level of fragmented GST-BHMT (GST-BHMT fragment), indicating an increase in basal autophagy, and treatment with 3-MA suppresses this increased basal autophagy.
(C) Immunoprecipitation with WAC antibody under glucose/glutamine (or energy) depletion followed by repletion (−/+) conditions shows WAC interaction with endogenous mTOR, Raptor, Pontin, Reptin, and TTI1, and with transfected FLAG-TEL2. IP with WAC(−) cells and probing for β-Catenin serve as negative controls. WCE, Whole cell extract.
(D), (E), and (F) Glucose/glutamine depletion (−) followed by repletion (−/+) induces an (D) increase in mTOR-dependent phosphorylation of S6K and AKT and (E) accumulation of p62, as well as (F) a decrease in the level of LC3-II. All these energy-dependent responses are inhibited by WAC knock down. Actin is the loading control for (D) and (E), and the non-specific band above WAC, likely corresponding to a protein unrelated to WAC as it showed up in almost all the WAC-KD cells, is the loading control for (F).
See also Figure S5.
To determine if the physical interactions between Wacky and mTOR pathway components identified through AP-MS in Drosophila are conserved, we performed co-immunoprecipitation (co-IP) experiments in HEK 293T cells. Under basal conditions, we were unable to co-IP mTOR and Raptor and the other mTOR pathway components using WAC antibody unless a cross-linking reagent, dithiobis (succinimidyl propionate) (DSP) (Smith et al., 2011; Zlatic et al., 2010), was included, indicating weak and/or transient physical association of WAC with these proteins (Figure S5C). WAC did not interact with a negative control, β-Catenin, a widely distributed protein. Furthermore, the mTOR pathway components were not detected in WAC pull down from WAC(−) cells, supporting our conclusion that these interactions are specific to WAC.
Since assembly of the larger TTT-Pontin/Reptin complex and its association with mTOR are energy dependent, and their physical interactions are more easily detectable following energetic stimulation (i.e., energy depletion followed by energy repletion) (Kim et al., 2013b), we repeated the experiments in energy repletion conditions. In this assay, cells are first starved of, then fed with glucose and glutamine prior to harvesting. Under this condition, without DSP, robust interactions between endogenous WAC and mTOR, Raptor, Pontin, Reptin, and TTI1, as well as transfected FLAG-tagged TEL2 are observed. These data provide compelling evidence that the physical interactions of WAC with mTORC1 and the TTT-Pontin/Reptin complex are conserved and potentially regulated by cellular energy status (Figure 5C).
To assess the significance of the physical interactions, we examined the effect of WAC depletion on mTOR activity under energy repletion conditions. Consistent with previous reports (Kim et al., 2013b), energy starvation results in mTOR inactivation, revealed by reduced levels of S6K and AKT phosphorylation (Figure 5D), and strong induction of autophagy, measured by increased LC3 lipidation (LC3-II) and reduced p62 (Figures 5E, 5F, S5D, and S5E). In contrast, energy-stimulated activation of mTOR is associated with increased levels of S6K and AKT phosphorylation (Figure 5D) and diminished levels of LC3-II as well as higher levels of p62 (Figures 5E, 5F, S5D, and S5E). However, depletion of WAC largely abolishes all of these mTOR-associated effects. Our observation that inclusion of the lysosomal inhibitor, Bafilomycin A1, similarly increases p62 and LC3-II levels in both controls and WAC(−) cells implies that loss of WAC does not affect autophagy flux and the observed effect of loss of WAC on p62 and LC3-II is primarily due to increased autophagic activity (Figures 5E, 5F, S5D, and S5E). Together, these findings demonstrate an important role of WAC in energy-induced mTOR activation.
WAC regulates mTORC1 activity by promoting the assembly of the TTT-Pontin/Reptin complex
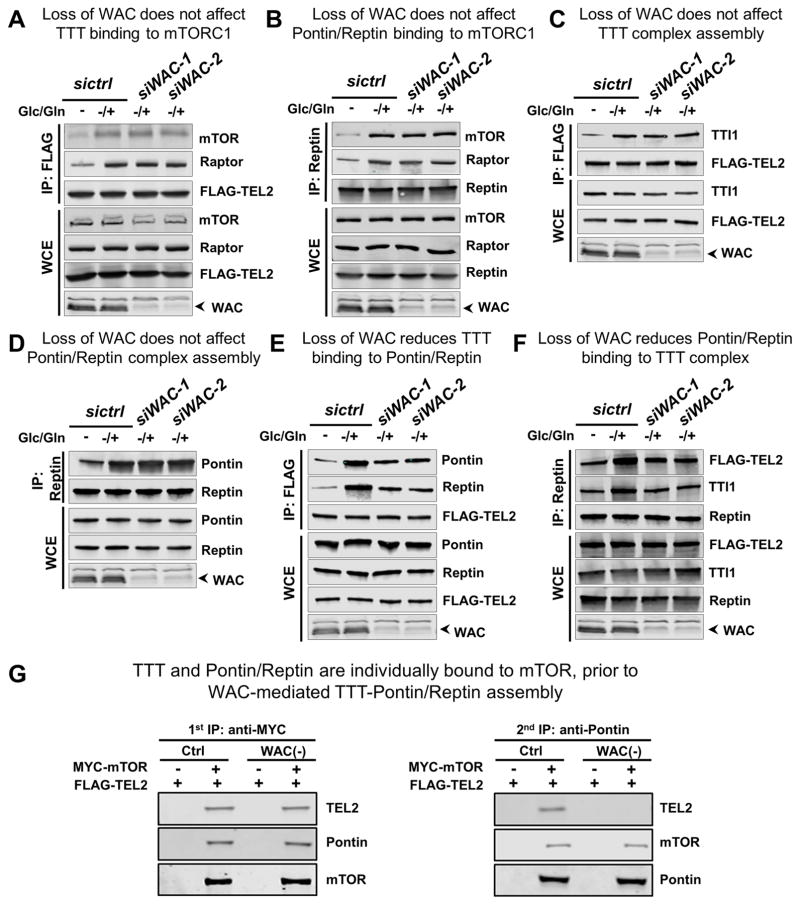
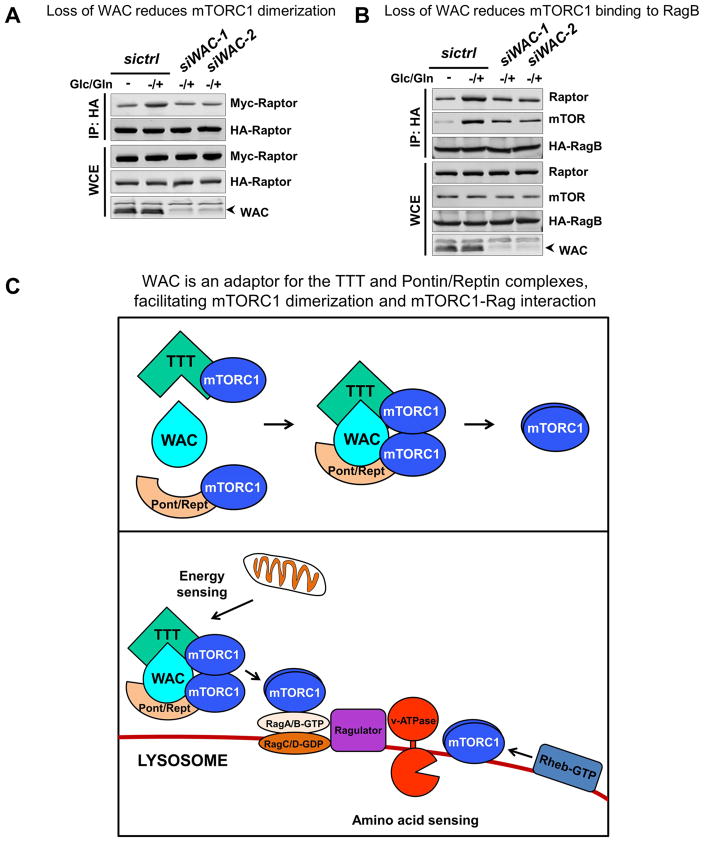
Our findings that WAC physically interacts with both mTORC1 and the TTT-Pontin/Reptin complex and is required for energy-dependent activation of mTORC1 (Figures 5 and S5), a process controlled by the TTT-Pontin/Reptin complex, implicate a functional link between WAC and this regulatory complex. To probe the underlying mechanism, we examined the role of WAC in the regulation of mTORC1 by the TTT-Pontin/Reptin complex. Consistent with previous observations (Kim et al., 2013b), energy stimulation promotes the association of mTORC1 with TTT and Pontin/Reptin (Figures 6A and 6B). Depletion of WAC, however, does not obviously disrupt mTORC1 interaction with TTT (Figure 6A) or with Pontin/Reptin (Figure 6B). Energy stimulation also leads to higher affinity among components of TTT (Figure 6C), an increased interaction between Pontin and Reptin (Figure 6D), as well as the assembly of the TTT-Pontin/Reptin complex (Figures 6E, 6F, and S6A). Interestingly, while loss of WAC does not affect the energy-dependent association among components of TTT or the interaction between Pontin and Reptin (Figures 6C and 6D), it markedly reduces the interaction between TTT and Pontin/Reptin (Figures 6E, 6F, S6A, S6B, and S6C). Further analysis suggests that TTT and Pontin/Reptin can each bind to mTOR separately, and WAC acts to promote their assembly into the larger TTT-Pontin/Reptin complex. In a sequential co-IP experiment, eluate from the initial pull-down with Myc-tagged mTOR was subjected to secondary co-IP with Pontin antibody, and while both TEL2 and mTOR are detected in the final pull-down with anti-Pontin in control cells, only mTOR is detected in WAC(−) cells (Figure 6G). These findings suggest that WAC regulates mTORC1 dimerization and mTORC1-Rag interaction, two interactions mediated by the TTT-Pontin/Reptin chaperone complex. To test this, we performed co-IP experiments with HA- and MYC-tagged Raptor to examine the dimerization status of mTORC1, and also with HA-tagged RagB and endogenous Raptor and mTOR to examine the association of Rag with mTORC1. Indeed, compared to control siRNA-treated cells, depletion of WAC significantly compromised the interaction between HA- and MYC-tagged Raptor (Figures 7A and S7A), and decreased the association of RagB with mTOR and Raptor (Figures 7B and S7B).
Figure 6. WAC promotes the assembly of the TTT-Pontin/Reptin complex.
Co-immunoprecipitation (co-IP) assays for HEK 293T cells treated with glucose/glutamine depletion (−) followed by glucose/glutamine repletion (−/+).
(A) and (B) Glucose/glutamine repletion enhances both the interaction (A) between FLAG-TEL2 and mTORC1 components, mTOR and Raptor, as shown by co-IP with anti-FLAG antibody, and (B) between Reptin and mTORC1 components, mTOR and Raptor, as shown by co-IP with anti Reptin. Both interactions are not affected by WAC knock down.
(C) and (D) Glucose/glutamine repletion enhances (C) the assembly of the TTT complex, as shown by co-IP with anti-FLAG antibody against FLAG-TEL2 and TTI1, as well as (D) the Pontin/Reptin interaction, as shown by co-IP with anti-Reptin. Both interactions are not affected by WAC knock down.
(E) and (F) Glucose/glutamine repletion enhances the binding between the TTT and Pontin/Reptin complexes, as shown by co-IP (E) with anti-FLAG-TEL2 or (F) with anti-endogenous Reptin. This interaction is reduced upon WAC knock down.
(G) Sequential immunoprecipitation shows that TTT and Pontin/Reptin can bind separately to mTOR prior to interaction with WAC. The first IP with anti-Myc shows that mTOR binds both Pontin and TEL2. The second IP with anti-Pontin (using eluate from the first IP) shows that, in WAC(−) cells, Pontin is able to bind mTOR, but not TEL2.
See also Figure S6.
Figure 7. WAC regulates mTORC1 activity by stabilizing the TTT-Pontin/Reptin complex.
(A) Glucose/glutamine repletion enhances mTORC1 dimerization, as shown by co-IP between HA-Raptor and Myc-Raptor. This dimerization is reduced upon WAC knock down.
(B) Glucose/glutamine repletion enhances Rag interaction with mTORC1 components, mTOR and Raptor, as shown by co-IP against HA-RagB. This interaction is reduced upon WAC knock down.
(C) WAC is an adaptor for the TTT and Pontin/Reptin complexes and facilitates their interaction, which is required for the energy-dependent increase in mTORC1 dimerization and mTORC1-Rag interaction.
See also Figure S7.
Taken together, these data show that WAC is necessary for the interaction between TTT and Pontin/Reptin, promoting mTORC1 activation and suppressing basal autophagy (Figure 7C).
DISCUSSION
WAC is a conserved regulator of mTOR activity
Regulation of metabolic activity in response to growth factors, nutrient availability, and energy levels is coordinated by mTORC1 through intricate mechanisms (Dibble and Manning, 2013; Kim et al., 2013a). Proper sensing of and response to changes in the environment of an organism are crucial in this delicate metabolic balance.
From a forward genetic screen to identify players in neuronal development, function, or maintenance in Drosophila, we identified Wacky, the Drosophila homolog of WAC. Loss of Wacky leads to animal lethality, aberrant development, neurodegeneration, and defective mTOR signaling. The latter results in developmental arrest, decreased cell size, increased basal autophagy, and diminished phosphorylation of mTOR targets (4E-BP, S6k and AKT). The requirement of WAC in mTOR regulation is conserved in mammalian cells, as loss of WAC leads to elevated basal autophagy and compromised mTOR activity in response to energy stimulation. Hence, WAC is a conserved positive regulator of mTOR.
WAC modulates the assembly of the TTT-Pontin/Reptin complex
mTOR activity is regulated through different mechanisms in response to specific stimuli (Figure S1A). The TTT and Pontin/Reptin complexes control energy-dependent activation of mTORC1 by chaperoning mTORC1 dimerization and by facilitating Rag-mediated lysosomal localization (Kim et al., 2013b). The dimerization of mTORC1 is an important, permissive step prior to the engagement of Rheb and Rag. Genetic analysis of wacky mutants supports its functional interaction with dTOR in a requisite step prior to Rheb- and Rag-mediated activation, as overexpression of dTOR but not active Rheb or Rag partially suppresses increased autophagy in wacky mutant clones. The physical interaction of Wacky with both dTOR and the Pontin/Reptin complex is consistent with the genetic data. In mammalian cells, WAC binds to mTOR and both Pontin/Reptin and TTT in an energy-dependent manner. In the absence of WAC, although both Pontin/Reptin and TTT form normally, the association between the two complexes is compromised. As a consequence, both mTORC1 dimerization and its interaction with Rag are affected, leading to diminished mTOR activity in WAC-depleted cells. However, overexpression of mTOR did not restore diminished mTORC1 activity in WAC-depleted HEK293T cells in response to energy stimulation (data not shown). The inability to partially suppress the phenotype in cells is most likely a reflection of evaluating mTOR activity in two very different experimental contexts: a chronic dTOR overexpression over many days in developing animals versus an assessment upon a 30-minute energy stimulation treatment in cultured WAC-knockdown cells. The TTT-Pontin/Reptin complex is known to regulate the maturation and stability of several PIKK family proteins, including mTOR, ATM, ATR and DNA-PKcs (Kim et al., 2013b). Consistent with a role for WAC in the assembly of the TTT-Pontin/Reptin complex, WAC knockdown in HEK293T cells leads to decreased levels of PIKK proteins (Figure S7C). Hence, WAC regulates mTOR by functioning as an adaptor to facilitate the assembly of the TTT-Pontin/Reptin complex in response to energy stimulation.
Little is known about how the assembly of the active TTT-Pontin/Reptin complex is regulated. Our study points to a role of WAC in this regulatory process. TTT and Pontin/Reptin have previously been isolated independently, but have also been observed in a single complex (Glatter et al., 2011; Hořejší et al., 2010; Hurov et al., 2010; Izumi et al., 2010; Kaizuka et al., 2010; Kim et al., 2013b; Nano and Houry, 2013; Takai et al., 2007, 2010). The association between TTT and Pontin/Reptin has been shown to be mediated by the phospho-dependent interaction between the Casein Kinase 2 (CK2)-phosphorylated TEL2 in the TTT complex and the PIH1D1 scaffold protein in the R2TP complex, which contains Pontin and Reptin (Hořejší et al., 2010). The stability of TTT and the larger TTT-Pontin/Reptin complex is energy-dependent. Energetic stress, through glucose/glutamine deprivation, leads to the disassembly of the TTT-Reptin/Pontin complex and reduced association between TEL2 and TTI1 (Kim et al., 2013b). WAC depletion does not affect the formation of the individual TTT and Pontin/Reptin complexes nor their interaction with mTORC1, but rather interferes with the association between the two complexes (Figure 6). This implies a role of WAC as an adaptor that recognizes the TTT and Pontin/Reptin complexes and stabilizes their interaction under energy-rich conditions. It remains to be determined how WAC regulates the assembly of the TTT-Pontin/Reptin complex. Does WAC directly interact with the TTT and Pontin/Reptin complexes, or indirectly through other intermediates such as mTORC1 components? Finally, are these interactions modulated through post-translational modifications in response to energy stimuli, given that WAC exists mainly in a tyrosine phosphorylated form (Xu and Arnaout, 2002)? Since the phenotypes of wacky mutants are weaker than dTOR mutants and can be partially suppressed by dTOR overexpression, it is likely that WAC plays a modulatory role to promote mTORC1 dimerization through the TTT-Pontin/Reptin complex.
WAC is a versatile adaptor involved in multiple cellular processes
Our finding that WAC regulates mTOR signaling by bringing TTT and Pontin/Reptin together further supports the role of WAC as a versatile adaptor in multiple cellular processes. WAC was originally identified as a WW domain-containing Adaptor with a Coiled-coil region (hence the name) that co-localizes with pre-mRNA splicing machinery (Xu and Arnaout, 2002). It stabilizes the E3 ligase RNF20/40 complex and mediates its interaction with RNA polymerase II, regulating transcription-coupled histone H2B ubiquitination (Zhang and Yu, 2011). WAC binds to VCIP135, mediating its association with the Golgi, and activating its deubiquitinating function to regulate Golgi biogenesis (Totsukawa et al., 2011). WAC also interacts with Beclin1 and UBQLN4, two proteins involved in autophagy regulation (Behrends et al., 2010; Lim et al., 2006; McKnight et al., 2012; N’Diaye et al., 2009). The endogenous interactors of WAC that we uncovered, including mTOR, Raptor, Rictor, Pontin, Reptin, TEL2, TTI1, and TTI2, all contain WW domain-binding motifs (as predicted from Salah et al., 2012) suggesting that these motifs are part of the structural basis of WAC-mediated regulation of the TTT-Pontin/Reptin and mTOR complexes.
WAC’s expanding list of binding partners might also explain WAC’s apparently opposite effects on autophagy regulation under different conditions. In our study, loss of Wacky/WAC results in increased basal autophagy both in flies (Figures 1C, S1C, 3A, and 3B) and in human cells (Figures 5A, 5B, S5A, and S5B). WAC depletion also compromised autophagy inhibition induced by energy stimulation (Figures 5E, 5F, S5D, and S5E). Hence, WAC acts as a positive regulator of mTOR and, indirectly, as a negative regulator of autophagy. In addition, WAC was isolated in an siRNA screen as a positive regulator of starvation-induced autophagy (McKnight et al., 2012). The latter is not inconsistent with our data as we observe a decrease in starvation-induced autophagy in wacky mutant clones in the fat body (Figure S3A) and WAC-depleted mammalian cells under starvation (data not shown). This differential regulation of autophagy might be achieved through WAC’s diverse binding partners. Its interaction with Beclin1 (Behrends et al., 2010; McKnight et al., 2012), a core component of the VPS34 complex whose activity is essential for autophagosome biogenesis (Russell et al., 2014; Wirth et al., 2013), may likely be important for this condition, especially since starvation robustly activates Beclin1 (Wei et al., 2008). Although further studies are needed to elucidate WAC’s diverse cellular activities, existing data support that, through its ability to interact with different proteins, WAC acts as a versatile adaptor in multiple cellular processes under various cellular contexts.
WAC in neurodevelopmental disorders
The data presented here show that WAC plays an important role in energy-mediated dimerization of mTORC1 through the TTT-Pontin/Reptin complex and that loss of wacky in Drosophila causes progressive neurodegeneration. Dysregulation of mTOR signaling has been implicated in autism and neurodegenerative disorders (Gkogkas et al., 2013; Lipton and Sahin, 2014; Tang et al., 2014 ), and missense mutations in the TTT complex component, TTI2, have been shown to cause intellectual disability (Langouët et al., 2013; Najmabadi et al., 2011). Interestingly, a cohort of patients was recently identified with de novo heterozygous loss of function mutations in WAC, exhibiting a syndrome characterized by developmental delay/intellectual disability, dysmorphic features, and hypotonia (DeSanto et al., 2015). Given the similar clinical manifestations that stem from loss of WAC and loss of TTI2, our findings linking WAC to mTOR signaling via the TTT-Pontin/Reptin complex may provide a potential mTOR-mediated basis for WAC-associated diseases.
EXPERIMENTAL PROCEDURES
Fly strains and clonal analysis, Transmission Electron Microscopy, Molecular cloning, Generation of transgenic flies, Immunostaining and imaging, Affinity Purification-Mass Spectrometry, Western blotting for Drosophila proteins, Cell culture and transfection, RNA interference, and Co-immunoprecipitation and western blotting for human proteins are described in Supplemental Experimental Procedures.
Electroretinogram Assay
ERGs were performed as previously described (Verstreken et al., 2003). Adult flies were glued to a glass slide, a recording probe was placed on the eye surface, and a reference probe was inserted into the thorax. 1-second light flashes were delivered using a halogen lamp and the response was recorded and analyzed using the WinWCP software.
GST-BHMT Assay
GST-BHMT assay was performed as described previously (Dennis and Mercer, 2009; Mercer et al., 2008; Rui et al., 2015). Briefly, HEK 293T cells transfected with pRK5-GST-BHMT and siRNAs against WAC were treated with 10 mM 3-MA (Sigma) or ATG7 siRNA, together with 11 μM leupeptin and 6 μM E-64d for 6 hours prior to immunoprecipitation. Whole cell lysate was centrifuged for 13,200 rpm at 4°C for 30 minutes and supernatant was incubated with glutathione agarose for 3 hours. Immunoprecipitated GST fusion proteins were analyzed by western blotting. GST antibody was used to detect both GST-BHMT and GST-BHMT-FRAG. GFP-Myc expression, driven by internal ribosomal binding sites in the pRK5-GST-BHMT plasmid, was detected by Myc antibody and served as normalization control, as reported in Mercer et al. (2008).
In vivo cross linking assays
HEK293T cells transfected as indicated were washed once with cold PBS, and incubated with 2 mM Dithiobis (succinimidyl propionate) (DSP, Life Technologies) freshly prepared in PBS for 2 hours on ice. Unreacted DSP was quenched by adding 1 M Tris pH 7.5 to a final concentration of 20 mM for 15 minutes. Cross-linked cells were then wash with cold PBS and lysed with 0.3% CHAPS lysis buffer as indicated in previous co-immunoprecipitation experiments. Anti-HA (Roche), anti-Myc (Santa Cruz), or anti-WAC (Millipore) was incubated with protein A/G agarose and pre-cleared cell lysates for 3 hours at 4°C. Co-immunoprecipitated proteins were uncrosslinked by adding dithiothreitol in sample buffer prior to western blotting analysis.
LC3 lipidation assay
Cells were harvested in 2% Triton X-100/PBS buffer containing protease inhibitors for maximum LC3-II extraction according to previous studies (Kimura et al., 2009). LC3-II and loading control, Actin, were detected by rabbit anti-LC3 antibody (MBL international) and mouse anti-Actin antibody (Chemicon), respectively. The level of LC3 lipidation was quantified as the ratio of the measured LC3-II to Actin levels. LC3-II synthesis (ΔLC3-II) was calculated as the difference between LC3-II levels at 2 hours and 4 hours after lysosomal inhibitor treatment.
Two-step co-immunoprecipitation
Two-step co-immunoprecipitation was performed according to procedures described in Rui et al., 2004. Briefly, six 60-mm dishes of HEK293T cells were transfected with FLAG-TEL2 alone or together with Myc-mTOR as indicated. At 36 hours after transfection, cells were lysed with CHAPS lysis buffer, sonicated briefly, and centrifuged. The supernatant was incubated with Myc antibody bound to Protein A/G-agarose beads for 2 hours at 4°C. Beads were washed with the lysis buffer, and the Myc-mTOR protein complex was eluted with 300 μl of lysis buffer containing 250 μg/ml Myc peptide (Sigma) for 2 hours at 4°C. The second immunoprecipitation was performed using 150 μl of eluate from the first immunoprecipitation and 350 μl of the lysis buffer containing 10 μl of -Pontin antibody followed by addition of Protein A/G-agarose beads.
Statistical analysis
Two-tailed Student’s t tests were used to analyze the data.
Supplementary Material
Highlights.
Loss of Drosophila WAC homolog, Wacky, affects neuronal development and maintenance
Wacky and WAC promote mTOR signaling in flies and in human cells
WAC promotes mTOR activation by mediating mTORC1 dimerization and Rag-association
WAC regulates energy-sensitive interaction between TTT and Pontin/Reptin complexes
Acknowledgments
We thank the Bloomington Drosophila Stock Center, Helmut Kramer, John Olson, and Utpal Banerjee for flies. We thank Adeel Jawaid, Vafa Bayat, Claire Haueter, Yu-Fei Chen, Clarissa Benitez, Xiao Shi, and Hsiang-Chih Lu for their support in the ERG screen of X chromosome mutants. We thank Yuchun He and Hongling Pan for injections to create transgenic lines. We thank Huda Zoghbi, Rodney Samaco, Ronald Richman and Jeehye Park for some cell culture experiments. We thank Sung Yun Jung and the BCM pathway Discovery Proteomics Core (P30 CA125123) for IP-Mass Spectrometry experiments. We thank Wan Hee Yoon, Karen Schulze, and Clayton Morrison for experimental advice and critical reading of the manuscript. Confocal microscopy at BCM is supported by the Intellectual and Developmental Disabilities Research Center (NIH 1U54 HD083092). Gabriela David-Morrison was supported by the Developmental Biology Program Training Grant (NICHD T32HD055200). This work was also supported by NIH R01-NS069880 (SZ), 1RC4GM096355-01 (HJB), and grants from the Robert A. and Renee E. Belfer Family Foundation, the Huffington Foundation, and Target ALS (HJB). Shinya Yamamoto is a fellow of the Jan and Dan Duncan Neurological Research Institute at Texas Children’s Hospital. HJB is an investigator of the Howard Hughes Medical Institute.
Footnotes
AUTHOR CONTRIBUTIONS
The authors have made the following declarations about their contributions: Conceived and designed the experiments: GD-M, ZX, SZ, HJB. Performed the experiments: GD-M, ZX, Y-NR, LD, ZZ. Analyzed the data: GD-M, ZX, SZ, HJB. Contributed reagents/materials/analysis tools: W-LC, MJ, SY, BX, KZ, HS. Wrote the paper: GD-M, SZ, HJB.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YY, Neufeld TP. Autophagy takes flight in Drosophila. FEBS Lett. 2010;584:1342–1349. doi: 10.1016/j.febslet.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis PB, Mercer CA. The GST-BHMT Assay and Related Assays for Autophagy. In: Klionsky DJ, editor. Methods in Enzymology. San Diego, California: Elsevier Inc; 2009. pp. 97–118. [DOI] [PubMed] [Google Scholar]
- DeSanto C, D’Aco K, Araujo GC, Shannon N, Study D, Vernon H, Rahrig A, Monaghan KG, Niu Z, Vitazka P, et al. WAC loss-of-function mutations cause a recognisable syndrome characterised by dysmorphic features, developmental delay and hypotonia and recapitulate 10p11.23 microdeletion syndrome. J Med Genet. 2015;52:754–761. doi: 10.1136/jmedgenet-2015-103069. [DOI] [PubMed] [Google Scholar]
- Dibble CC, Manning BD. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat Cell Biol. 2013;15:555–564. doi: 10.1038/ncb2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskelinen E-L. Fine structure of the autophagosome. In: Deretic V, editor. Autophagosome and Phagosome. Totowa, New Jersey: Humana Press; 2008. pp. 11–28. [Google Scholar]
- Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya N, Kanazawa T, Fujimura S, Ueno T, Kominami E, Kadowaki M. Leupeptin-induced appearance of partial fragment of betaine homocysteine methyltransferase during autophagic maturation in rat hepatocytes. J Biochem. 2001;129:313–320. doi: 10.1093/oxfordjournals.jbchem.a002859. [DOI] [PubMed] [Google Scholar]
- Gkogkas CG, Khoutorsky A, Ran I, Rampakakis E, Nevarko T, Weatherill DB, Vasuta C, Yee S, Truitt M, Dallaire P, et al. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature. 2013;493:371–377. doi: 10.1038/nature11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatter T, Schittenhelm RB, Rinner O, Roguska K, Wepf A, Jünger MA, Köhler K, Jevtov I, Choi H, Schmidt A, et al. Modularity and hormone sensitivity of the Drosophila melanogaster insulin receptor/target of rapamycin interaction proteome. Mol Syst Biol. 2011;7:1–15. doi: 10.1038/msb.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK Phosphorylation of Raptor Mediates a Metabolic Checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haelterman NA, Jiang L, Li Y, Bayat V, Sandoval H, Ugur B, Tan KL, Zhang K, Bei D, Xiong B, et al. Large-scale identification of chemically induced mutations in Drosophila melanogaster. Genome Res. 2014;24:1707–1718. doi: 10.1101/gr.174615.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig KM, Neufeld TP. Inhibition of cellular growth and proliferation by dTOR overexpression in Drosophila. Genesis. 2002;34:107–110. doi: 10.1002/gene.10139. [DOI] [PubMed] [Google Scholar]
- Hořejší Z, Takai H, Adelman CA, Collis SJ, Flynn H, Maslen S, Skehel JM, de Lange T, Boulton SJ. CK2 phospho-dependent binding of R2TP complex to TEL2 is essential for mTOR and SMG1 stability. Mol Cell. 2010;39:839–850. doi: 10.1016/j.molcel.2010.08.037. [DOI] [PubMed] [Google Scholar]
- Huen J, Kakihara Y, Ugwu F, Cheung KLY, Ortega J, Houry WA. Rvb1-Rvb2: essential ATP-dependent helicases for critical complexes. Biochem Cell Biol. 2010;88:29–40. doi: 10.1139/o09-122. [DOI] [PubMed] [Google Scholar]
- Hurov KE, Cotta-Ramusino C, Elledge SJ. A genetic screen identifies the Triple T complex required for DNA damage signaling and ATM and ATR stability. Genes Dev. 2010;24:1939–1950. doi: 10.1101/gad.1934210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Xu T, Guan KL. Rheb GTpase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003a;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003b;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- Izumi N, Yamashita A, Iwamatsu A, Kurata R, Nakamura H, Saari B, Hirano H, Anderson P, Ohno S. AAA+ proteins RUVBL1 and RUVBL2 coordinate PIKK activity and function in nonsense-mediated mRNA decay. Sci Signal. 2010;3:ra27. doi: 10.1126/scisignal.2000468. [DOI] [PubMed] [Google Scholar]
- Jaiswal M, Haelterman NA, Sandoval H, Xiong B, Donti T, Kalsotra A, Yamamoto S, Cooper TA, Graham BH, Bellen HJ. Impaired Mitochondrial Energy Production Causes Light-Induced Photoreceptor Degeneration Independent of Oxidative Stress. PLoS Biol. 2015;13:e1002197. doi: 10.1371/journal.pbio.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaizuka T, Hara T, Oshiro N, Kikkawa U, Yonezawa K, Takehana K, Iemura SI, Natsume T, Mizushima N. Tti1 and Tel2 are critical factors in mammalian target of rapamycin complex assembly. J Biol Chem. 2010;285:20109–20116. doi: 10.1074/jbc.M110.121699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Guan K. mTOR3: a pharmacologic target for autophagy regulation. J Clin Invest. 2015;125:25–32. doi: 10.1172/JCI73939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SG, Buel GR, Blenis J. Nutrient regulation of the mTOR Complex 1 signaling pathway. Mol Cells. 2013a;35:463–473. doi: 10.1007/s10059-013-0138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SG, Hoffman GR, Poulogiannis G, Buel GR, Jang YJ, Lee KW, Kim BY, Erikson RL, Cantley LC, Choo AY, et al. Metabolic Stress Controls mTORC1 Lysosomal Localization and Dimerization by Regulating the TTT-RUVBL1/2 Complex. Mol Cell. 2013b;49:172–185. doi: 10.1016/j.molcel.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Fujita N, Noda T, Yoshimori T. Monitoring autophagy in mammalian cultured cells through the dynamics of LC3. Methods Enzymol. 2009;452:1–12. doi: 10.1016/S0076-6879(08)03601-X. [DOI] [PubMed] [Google Scholar]
- Klionsky D, Agholme L, Agnello M, Agostinis P, Aguirre-ghiso JA, Ahn HJ, Ait-mohamed O, Brown EJ, Brumell JH, Brunetti-pierri N, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFever L, Feoktistov A, Hsu HJ, Drummond-Barbosa D. Specific roles of Target of rapamycin in the control of stem cells and their progeny in the Drosophila ovary. Development. 2010;137:2451–2451. doi: 10.1242/dev.050351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langouët M, Saadi A, Rieunier G, Moutton S, Siquier-Pernet K, Fernet M, Nitschke P, Munnich A, Stern MH, Chaouch M, et al. Mutation in TTI2 reveals a role for triple T complex in human brain development. Hum Mutat. 2013;34:1472–1476. doi: 10.1002/humu.22399. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. MTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Hao T, Shaw C, Patel AJ, Szabó G, Rual JF, Fisk CJ, Li N, Smolyar A, Hill DE, et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- Lipton JO, Sahin M. The Neurology of mTOR. Neuron. 2014;84:275–291. doi: 10.1016/j.neuron.2014.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lullmann-Rauch R. History and morphology of the lysosome. In: Saftig P, editor. Lysosomes. Georgetown, Texas: Landes Bioscience/Eurekah.com; 2005. pp. 1–16. [Google Scholar]
- McKnight NC, Jefferies HBJ, Alemu EA, Saunders RE, Howell M, Johansen T, Tooze SA. Genome-wide siRNA screen reveals amino acid starvation-induced autophagy requires SCOC and WAC. EMBO J. 2012;31:1931–1946. doi: 10.1038/emboj.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer CA, Kaliappan A, Dennis PB. Macroautophagy-dependent, intralysosomal cleavage of a betaine homocysteine methyltransferase fusion protein requires stable multimerization. Autophagy. 2008;4:185–194. doi: 10.4161/auto.5275. [DOI] [PubMed] [Google Scholar]
- N’Diaye EN, Kajihara KK, Hsieh I, Morisaki H, Debnath J, Brown EJ. PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep. 2009;10:173–179. doi: 10.1038/embor.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najmabadi H, Hu H, Garshasbi M, Zemojtel T, Abedini SS, Chen W, Hosseini M, Behjati F, Haas S, Jamali P, et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478:57–63. doi: 10.1038/nature10423. [DOI] [PubMed] [Google Scholar]
- Nano N, Houry WA. Chaperone-like activity of the AAA+ proteins Rvb1 and Rvb2 in the assembly of various complexes. Philos Trans R Soc Lond B Biol Sci. 2013;368:20110399. doi: 10.1098/rstb.2011.0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld TP, Baehrecke EH. Eating on the fly: Function and regulation of autophagy during cell growth, survival and death in Drosophila. Autophagy. 2008;4:557–562. doi: 10.4161/auto.5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumüller RA, Wirtz-Peitz F, Lee S, Kwon Y, Buckner M, Hoskins RA, Venken KJT, Bellen HJ, Mohr SE, Perrimon N. Stringent analysis of gene function and protein-protein interactions using fluorescently tagged genes. Genetics. 2012;190:931–940. doi: 10.1534/genetics.111.136465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezis IP, Lamark T, Velentzas AD, Rusten TE, Bjørkøy G, Johansen T, Papassideri S, Stravopodis DJ, Margaritis LH, Stenmark H, et al. Cell death during Drosophila melanogaster early oogenesis is mediated through autophagy. Autophagy. 2009;5:298–302. doi: 10.4161/auto.5.3.7454. [DOI] [PubMed] [Google Scholar]
- Oldham S, Montagne J, Radimerski T, Thomas G, Hafen E. Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev. 2000;14:2689–2694. doi: 10.1101/gad.845700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel PH, Tamanoi F. Increased Rheb-TOR signaling enhances sensitivity of the whole organism to oxidative stress. J Cell Sci. 2006;119:4285–4292. doi: 10.1242/jcs.03199. [DOI] [PubMed] [Google Scholar]
- Pulipparacharuvil S, Akbar MA, Ray S, Sevrioukov EA, Haberman AS, Rohrer J, Krämer H. Drosophila Vps16A is required for trafficking to lysosomes and biogenesis of pigment granules. J Cell Sci. 2005;118:3663–3673. doi: 10.1242/jcs.02502. [DOI] [PubMed] [Google Scholar]
- Rui Y, Xu Z, Lin S, Li Q, Rui H, Luo W, Zhou HM, Cheung PY, Wu Z, Ye Z, et al. Axin stimulates p53 functions by activation of HIPK2 kinase through multimeric complex formation. EMBO J. 2004;23:4583–4594. doi: 10.1038/sj.emboj.7600475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui YN, Xu Z, Patel B, Chen Z, Chen D, Tito A, David G, Sun Y, Stimming EF, Bellen HJ, et al. Huntingtin functions as a scaffold for selective macroautophagy. Nat Cell Biol. 2015;17:262–275. doi: 10.1038/ncb3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RC, Yuan HX, Guan KL. Autophagy regulation by nutrient signaling. Cell Res. 2014;24:42–57. doi: 10.1038/cr.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten TE, Lindmo K, Juhász G, Sass M, Seglen PO, Brech A, Stenmark H. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell. 2004;7:179–192. doi: 10.1016/j.devcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Salah Z, Alian A, Aqeilan RI. WW domain-containing proteins: retrospectives and the future. Front Biosci. 2012;17:331–348. doi: 10.2741/3930. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval H, Yao CK, Chen K, Jaiswal M, Donti T, Lin YQ, Bayat V, Xiong B, Zhang K, David G, et al. Mitochondrial fusion but not fission regulates larval growth and synaptic development through steroid hormone production. Elife. 2014;3:1–23. doi: 10.7554/eLife.03558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Smith AL, Friedman DB, Yu H, Carnahan RH, Reynolds AB. ReCLIP (reversible cross-link immuno-precipitation): an efficient method for interrogation of labile protein complexes. PLoS One. 2011;6:e16206. doi: 10.1371/journal.pone.0016206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H, Wang RC, Takai KK, Yang H, de Lange T. Tel2 Regulates the Stability of PI3K-Related Protein Kinases. Cell. 2007;131:1248–1259. doi: 10.1016/j.cell.2007.10.052. [DOI] [PubMed] [Google Scholar]
- Takai H, Xie Y, De Lange T, Pavletich NP. Tel2 structure and function in the Hsp90-dependent maturation of mTOR and ATR complexes. Genes Dev. 2010;24:2019–2030. doi: 10.1101/gad.1956410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Gudsnuk K, Kuo SH, Cotrina ML, Rosoklija G, Sosunov A, Sonders MS, Kanter E, Castagna C, Yamamoto A, et al. Loss of mTOR-Dependent Macroautophagy Causes Autistic-like Synaptic Pruning Deficits. Neuron. 2014;83:1131–1143. doi: 10.1016/j.neuron.2014.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous Sclerosis Complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- Tian X, Gala U, Zhang Y, Shang W, Nagarkar Jaiswal S, di Ronza A, Jaiswal M, Yamamoto S, Sandoval H, Duraine L, et al. A Voltage-Gated Calcium Channel Regulates Lysosomal Fusion with Endosomes and Autophagosomes and Is Required for Neuronal Homeostasis. PLOS Biol. 2015;13:e1002103. doi: 10.1371/journal.pbio.1002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totsukawa G, Kaneko Y, Uchiyama K, Toh H, Tamura K, Kondo H. VCIP135 deubiquitinase and its binding protein, WAC, in p97ATPase-mediated membrane fusion. EMBO J. 2011;30:3581–3593. doi: 10.1038/emboj.2011.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno T, Ishidoh K, Mineki R, Tanida I, Murayama K, Kadowaki M, Kominami E. Autolysosomal membrane-associated betaine homocysteine methyltransferase. Limited degradation fragment of a sequestered cytosolic enzyme monitoring autophagy. J Biol Chem. 1999;274:15222–15229. doi: 10.1074/jbc.274.21.15222. [DOI] [PubMed] [Google Scholar]
- Verstreken P, Koh TW, Schulze KL, Zhai RG, Hiesinger PR, Zhou Y, Mehta SQ, Cao Y, Roos J, Bellen HJ. Synaptojanin is recruited by endophilin to promote synaptic vesicle uncoating. Neuron. 2003;40:733–748. doi: 10.1016/s0896-6273(03)00644-5. [DOI] [PubMed] [Google Scholar]
- Wang S, Tan KL, Agosto MA, Xiong B, Yamamoto S, Sandoval H, Jaiswal M, Bayat V, Zhang K, Charng WL, et al. The Retromer Complex Is Required for Rhodopsin Recycling and Its Loss Leads to Photoreceptor Degeneration. PLoS Biol. 2014;12:e1001847. doi: 10.1371/journal.pbio.1001847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Blumhagen R, Lao U, Kuo Y, Edgar BA. LST8 Regulates Cell Growth via Target-of-Rapamycin Complex 2 (TORC2) Mol Cell Biol. 2012;32:2203–2213. doi: 10.1128/MCB.06474-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-Mediated Phosphorylation of Bcl-2 Regulates Starvation-Induced Autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth M, Joachim J, Tooze SA. Autophagosome formation-The role of ULK1 and Beclin1-PI3KC3 complexes in setting the stage. Semin Cancer Biol. 2013;23:301–309. doi: 10.1016/j.semcancer.2013.05.007. [DOI] [PubMed] [Google Scholar]
- Xiong B, Bayat V, Jaiswal M, Zhang K, Sandoval H, Charng WL, Li T, David G, Duraine L, Lin YQ, et al. Crag Is a GEF for Rab11 Required for Rhodopsin Trafficking and Maintenance of Adult Photoreceptor Cells. PLoS Biol. 2012;10:e1001438. doi: 10.1371/journal.pbio.1001438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu GM, Arnaout MA. WAC, a novel WW domain-containing adapter with a coiled-coil region, is colocalized with splicing factor SC35. Genomics. 2002;79:87–94. doi: 10.1006/geno.2001.6684. [DOI] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Yamamoto S, Jaiswal M, Charng W, Gambin T, Karaca E, Mirzaa G, Wiszniewski W, Sandoval H, Haelterman NA, Xiong B, et al. A Drosophila Genetic Resource of Mutants to Study Mechanisms Underlying Human Genetic Diseases. Cell. 2014;159:200–214. doi: 10.1016/j.cell.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Yu X. WAC, a functional partner of RNF20/40, regulates histone H2B ubiquitination and gene transcription. Mol Cell. 2011;41:384–397. doi: 10.1016/j.molcel.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Stallock JP, Ng JC, Reinhard C, Neufeld TP. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 2000;14:2712–2724. doi: 10.1101/gad.835000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Li Z, Jaiswal M, Bayat V, Xiong B, Sandoval H, Charng WL, David G, Haueter C, Yamamoto S, et al. The C8ORF38 homologue Sicily is a cytosolic chaperone for a mitochondrial complex I subunit. J Cell Biol. 2013;200:807–820. doi: 10.1083/jcb.201208033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirin J, Perrimon N. Drosophila as a model system to study autophagy. Semin Immunopathol. 2010;32:363–372. doi: 10.1007/s00281-010-0223-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatic SA, Ryder PV, Salazar G, Faundez V. Isolation of Labile Multi-protein Complexes by in vivo Controlled Cellular Cross-Linking and Immuno-magnetic Affinity Chromatography. J Vis Exp. 2010 doi: 10.3791/1855. 1855 [pii], 1810.3791/1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 Senses Lysosomal Amino Acids Through an Inside-Out Mechanism That Requires the Vacuolar H+-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.