Fig. 2. Characterization of αHL-GroES chimera proteins.
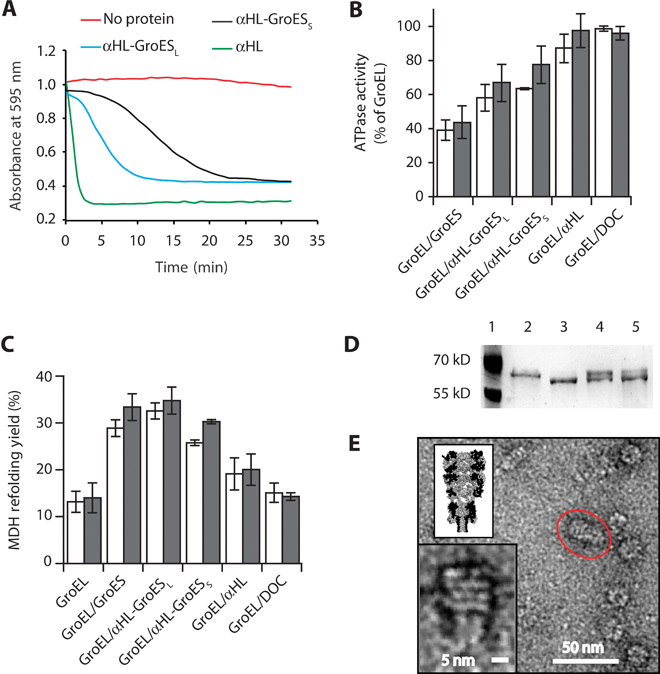
(A) Typical traces for the hemolytic assay showing the pore-forming activity of nanopore proteins. Each line shows the reduction in absorbance for monomeric nanopore proteins: αHL is in green, αHL-GroESL is in blue, and αHL-GroESS is in black. The red line shows a control experiment where no protein was used. Proteins are added to a 0.0625 μM final concentration and were incubated with a solution of diluted rabbit red blood cells in 10 mM Mops and 150 mM NaCl (pH 7.4) containing bovine serum albumin (1 mg/ml). The pore-forming activity of the αHL proteins is shown by the decrease in the absorbance at 595 nm due to the lysis of the rabbit red blood cells. (B) ATPase activity of GroEL (50 nM) in the presence of GroES proteins (200 nM) and/or the surfactant used to solubilize the nanopores [deoxycholate (DOC), 0.125 mM] in 50 mM KCl (white bars) or 1 M KCl (gray bars). The ATPase reaction was started by adding ATP to the reaction buffer containing GroES and GroEL proteins. The values are shown in table S2. (C) Refolding assay assisted by GroES proteins in 50 mM KCl (white bars) or 1 M KCl (gray bars). Unfolded MDH (2 μM) was preincubated with GroEL (50 nM) in the refolding buffer (no ATP) before the addition of ATP (2 mM) and co-chaperonin proteins (200 nM). Control experiments with DOC and αHL indicated that the GroEL-assisted refolding is mediated by the grafted loops in αHL-GroESL. Refolding yields were normalized by the spontaneous refolding of MDH. The values are shown in table S3. (D) Proteinase K protection assay. GroEL (0.2 μM), preincubated with GroES or αHL-GroESL (1 μM) in lanes 4 and 5, respectively, was treated with proteinase K (25 μg/ml) in the presence of ATP (1 mM) for 20 min before loading into polyacrylamide SDS gels. Lane 1, protein ladder. Lane 2, undigested GroEL (58 kD). Lane 3, GroEL after incubation with proteinase K (~56 kD). Lane 4, GroEL digestion protected by GroES. Lane 5, GroEL digestion protected by αHL-GroESL. (E) Negative-stained EM image of GroEL-398 bound to αHL-GroESL formed by preincubating GroEL-398 (0.5 μM) with αHL-GroESL (1 μM) for 20 min before applying a 100-fold dilution to negatively stained EM grids. The insets show the magnification of the circled complex (bottom) and a scaled surface representation of the αHL-GroESL:GroEL complex (top). Experiments and dilutions in (B) and (C) were carried out in 50 mM tris-HCl (pH 7.5), 50 mM KCl, 1 mM ATP, and 5 mM MgCl2. Errors are quoted as SD.
