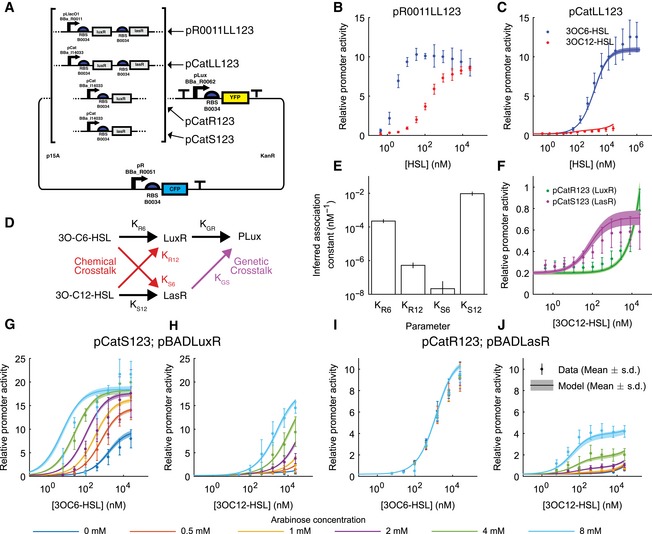
-
A
Ratiometric reporter constructs express eYFP under the control of pLux (Bba_R0062) and eCFP under pR (Bba_R0051) but differ in which receiver proteins are expressed and the strength of their expression (described in
Appendix Table S1). Strong expression is driven by pLlacO1 (Bba_R0011) and weak expression by pCat (Bba_I14033).
-
B
The relative activity of pLux in the presence of strong expression of both receiver proteins (plasmid pR0011LL123) as a function of 3OC6HSL concentration (blue points) and 3OC12HSL concentration (red points).
-
C
The relative activity of pLux in the presence of weak expression of both receiver proteins (plasmid pCatLL123) as a function of 3OC6HSL concentration (blue points) and 3OC12HSL concentration (red points).
-
D
Crosstalk can occur due to receiver protein binding noncognate HSL (chemical crosstalk) or due to signal‐bound receiver activating transcription at a noncognate promoter (genetic crosstalk).
-
E
Inferred association constants for LuxR or LasR with 3OC6HSL or 3OC12HSL
-
F
The relative activity of pLux in the presence of weak expression of LuxR (plasmid pCatR123, green points and line) or weak expression of LasR (plasmid pCatS123, magenta points and line) as a function of 3OC12HSL concentration.
-
G–J
The relative activity of pLux in the presence of weak expression of LasR and inducible expression of LuxR (G, H, plasmids pCatS123 and pBADLuxR) or weak expression of LuxR and inducible expression of LasR (I, J, plasmids pCatR123 and pBADLasR) as a function of 3OC6HSL concentration (G, I) or 3OC12HSL concentration (H, J). Inducible expression was varied via arabinose concentration as indicated by the color code.
Data information: (B, C, F–I) Relative promoter activity (ρ = deYFP/deCFP) with respect to the reference pR is reported as a function of HSL concentration. Points indicate the mean of three replicates and error bars indicate the standard deviation while lines and shading indicate the mean and standard deviation of the best‐fit models, respectively.
Source data are available online for this figure.