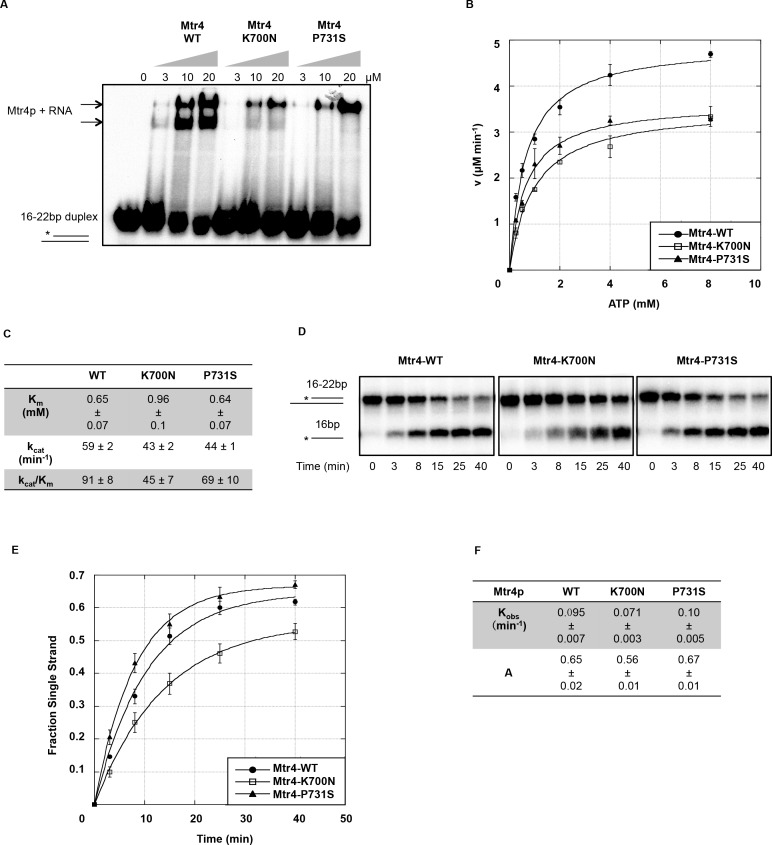
Fig 5. Mtr4p Arch Domain Mutants Exhibit Defects in RNA Binding and ATP Hydrolysis, but showed marginal defects in RNA unwinding in vitro.
A) RNA binding of Mtr4p and Mtr4 arch domain mutants by EMSA. Concentrations of Mtr4p used in each reaction are listed above the lanes. The arrows indicate the position(s) of Mtr4p-RNA complexes. The duplex RNA structure (R1-4) is shown as a cartoon, indicating the migration of unbound duplex RNA, with the 32P radiolabeled strand marked with an asterisk. B) RNA-dependent ATP hydrolysis activity of Mtr4p and Mtr4p arch domain mutants. ATPase assays were conducted with purified recombinant wild type or mutant Mtr4p at different ATP concentrations (0.25 mM, 0.5 mM, 1.0 mM, 2.0 mM, 4.0 mM, 8.0 mM). Near saturating levels of Escherichia coli total tRNA (Roche) (9 μM) was used in each reaction to initiate Mtr4p’s ATP hydrolysis activity. Rate plots of ATP hydrolysis versus ATP concentration done in triplicate are shown as best fit plots from the Michaelis-Menton equation. C) Kinetic studies to assess the efficiency of Mtr4p arch domain mutants to hydrolyze ATP in the presence of saturating RNA. The. Km represents the dissociation constant of Mtr4p for ATP. kcat/Km provides measurement of Mtr4p ATP hydrolysis efficiency. D) Representative native-PAGE of unwinding reactions where 50 nM of wild type or mutant Mtr4 protein was used in each RNA-unwinding reaction at 30°C for the times shown. Cartoons on the left indicate the migration distance of radiolabeled (*) 16 base single stranded RNA and the 22/16 duplex RNA E) Time courses of unwinding reactions for wild-type and mutant Mtr4 proteins done in triplicate are shown as plots where the proportion of single stranded RNA is plotted against reaction time and fitted to a first-order reaction. F) Shown are the kinetic parameters of RNA duplex unwinding by wild-type and arch domain mutant Mtr4p, where the reaction amplitude (A) represents the fraction of unwound single-stranded RNA, and the Kobs or rate constant that indicates enzyme efficiency of unwinding.