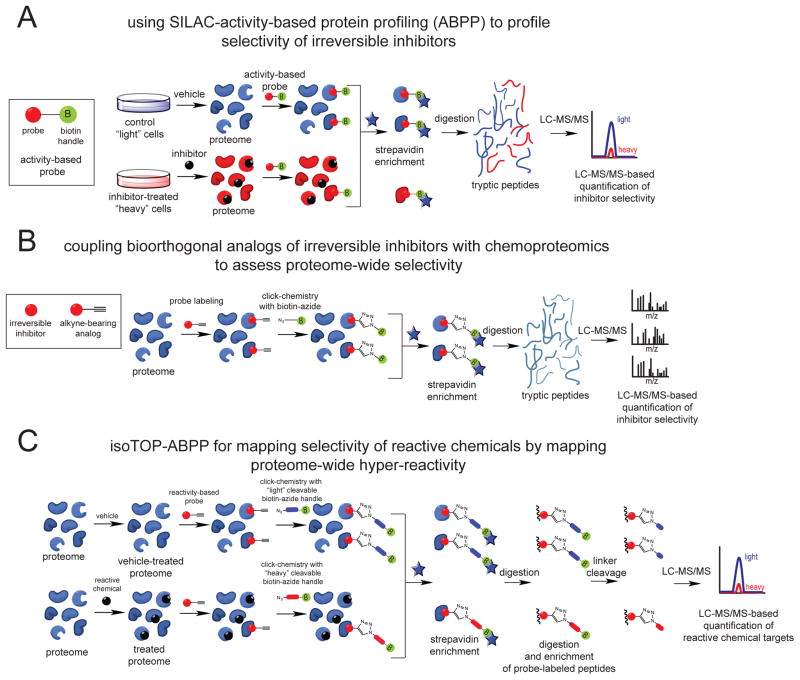
Figure 2. Chemoproteomic platforms for assessing proteome-wide targets of irreversibly-acting chemicals.
(A) SILAC-ABPP uses active-site directed chemical probes to assess the functional state of large numbers of enzymes directly in complex proteomes. Small-molecule inhibitors can be competed against the binding of activity-based probes to enzymes to assess enzyme class-wide selectivity. Cells can be labeled with light or heavy isotopic amino acids for quantitative proteomic analysis. (B) Analogs of these inhibitors bearing a bioorthogonal handle (e.g. alkyne) can be used to assess proteome-wide selectivity of small-molecule inhibitors using chemoproteomic approaches. (C) Isotopic Tandem Orthogonal Proteolysis-ABPP (isoTOP-ABPP) can be used to map hyper-reactive and functional sites across the proteome using reactivity-based chemical probes bearing bioorthogonal handles (e.g. alkyne). Reactive electrophiles can be competed against probe binding to hyper-reactive sites to map protein targets of these reactive agents. Probe-labeled peptides can be identified through subsequent appending of a biotin-azide analytical handle bearing a TEV protease recognition sequence and heavy or light isotopic valine tag using copper-catalyzed click chemistry. Upon mixing control and treated proteomes, probe-labeled proteins can be avidin-enriched, tryptically digested, and probe-labeled peptides can be subsequently enriched and released by TEV protease for subsequent quantitative proteomic analysis.