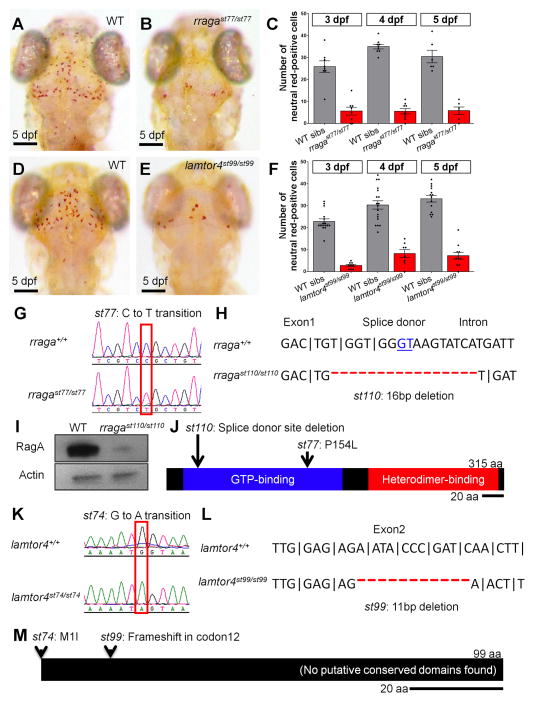
Figure 1. Mutational analysis demonstrates that rraga and lamtor4 are essential for microglia development.
(A–F) Comparison of microglia numbers in rragast77/st77 mutants, lamtor4st99/st99 mutants, and wildtype siblings. (A, B, D, E) Neutral red stained larvae of the indicated genotypes at 5 dpf. Dorsal views, scale bar = 50 μm. (C, F) Quantification of neutral red stained microglia in rraga (C) and lamtor4 (F) mutants at 3 dpf, 4 dpf and 5 dpf. (G–J) Molecular analysis of mutations in rraga. (G) Sequence chromatograms show point mutation in rragast77 mutation. (H) Sequence deleted in rragast110 mutation, showing exon-intron boundary and deleted splice donor. (I) Immunoblot showing pronounced reduction of RagA protein in rragast77/st77 mutants at 5 dpf. Actin is shown as a loading control. (J) Schematic of RagA protein, showing conserved domains and positions of the mutant lesions. (K–M) Molecular analysis of mutations in lamtor4. (K) Sequence chromatograms show point mutation in lamtor4st74 mutation, which changes the ATG initiation codon to ATA. (L) Sequence deleted in lamtor4st99 mutation, showing the reading frame in wildtype and the alteration caused by the 11 bp deletion in the mutation. (M) Schematic of the Lamtor4 protein, showing positions of the mutant lesions. Larvae shown in A, B, D, and E were genotyped by PCR after photography.