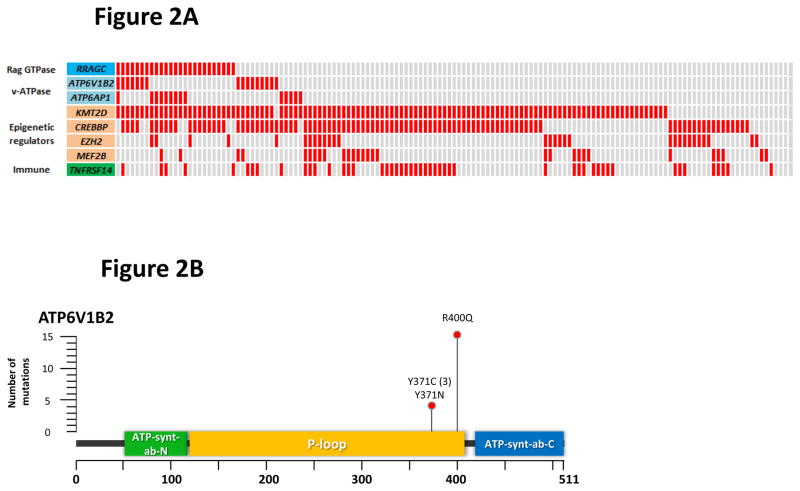
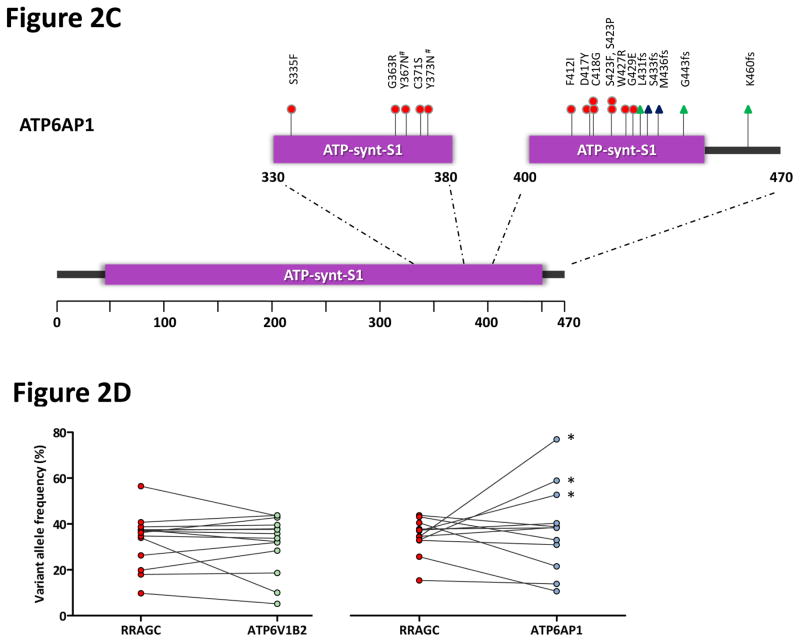
Figure 2.
Frequent and co-occurring mutations in ATP6V1B2 and ATP6AP1. (a) The heatmap shows the distribution of mutations in RRAGC, ATP6V1B2, ATP6AP1 and other known FL-associated genes in 141 FL cases. Each column represents an individual case and each row denotes a specific gene. Red indicates the presence of mutations, and light grey indicates the absence. (b) Schema of the protein domain and locations of the identified ATP6V1B2 mutations (NCBI protein reference sequence: NP_001684.2). (c) Schema of the protein domain and locations of the identified ATP6AP1 mutations (NCBI protein reference sequence: NP_001174.2). Red circles represent missense mutations, blue triangles are in-frame indels and green triangles are out-of-frame indels. The ‘#’ indicates mutations occurring in the same case. (d) Comparison of the allele frequencies in 26 co-mutated (RRAGC vs. ATP6V1B2 and RRAGC vs. ATP6AP1) samples (comprising 15 cases, some with multiple biopsies). Male cases are marked with an asterisk (*) and demonstrate expected increases in allelic frequencies of ATP6AP1 mutations as the gene locus resides on the X chromosome.