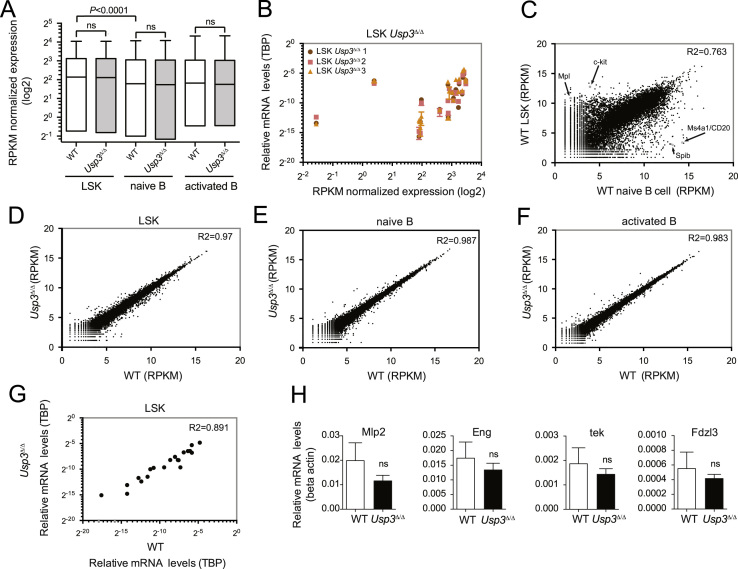
Fig. 1.
Analysis and validation of RNA-seq data from WT and Usp3Δ/Δ mice. (A) Expression levels of WT and Usp3Δ/Δ freshly isolated LSK and naïve B cells, and in vitro lipopolysaccharide (LPS) stimulated B cells (activated B cell). Statistical significant difference between the transcriptional profile of LSK versus naïve B cells is shown: ****P<0.0001. RPKM Reads Per Kilobase of transcript per Million mapped reads. (B) Expression of a set of 19 genes (Supplementary Table S1) was assessed by qRT-PCR in three independent LSK mRNA per each genotype. Results for the Usp3Δ/Δ genotype are shown. qRT-PCR and the RNA-seq normalized expression data for these genes in Usp3Δ/Δ samples had a good linear relationship, validating the RNAseq analysis. Pearson coefficient of LSK RNA-seq Usp3Δ/Δ compared with LSK qRT-PCR of: Usp3Δ/Δ 1, r=0.4853; Usp3Δ/Δ 2, r=0.4777; Usp3Δ/Δ 3, r=0.4964. *P<0.05. (C) Comparison of normalized gene expression data for WT LSK versus WT naïve B cells of one representative experiment. Pearson coefficient r=0.874. Distinct LSK-specific and B-cell specific expressed genes are recognizable. Among the LSK-specific genes, the Mpl receptor [6] and the Kit receptor [7] are indicated (gray dots). MS4A1/CD20 and Spi-B transcription factor are genes specifically expressed in B cells (gray dots). (D–F) Comparison of normalized gene expression data for Usp3Δ/Δ versus WT LSK (Pearson coefficient r=0.986, R2 coefficient=0.9738) (D), naïve B cell (Pearson coefficient r=0.974, R2=0.987) (E) and LPS activated B cell (Pearson coefficient r=0.991, R2 coefficient=0.983) (F). One representative experiment out of two is shown. (G) qRT-PCR analysis of a set of 19 genes in Usp3Δ/Δ and WT LSK (Supplementary Table S1). Pearson coefficient r=0.9443; R2 coefficient=0.891. Single qRT-PCR results for a subset of HSC-specific genes, Mpl2, Eng, Tek and Fdzl3 [8], is shown in (H). Data are from three independent LSK mRNA per genotype. Data are means±SEM. (A), (C–F) Two independent experiments were performed. For each experiment, pooled cells from 4 individual mice/genotype (8 weeks old) were analyzed by RNA-seq. Cells were: LSK, FACS-sorted LSKs from bone marrow; naïve B, FACS-sorted B cells from spleen (CD19+); activated B, B cells harvested and FACS sorted after 4 days stimulation with LPS in culture. (B), (G) qRT-PCR was performed on mRNA from FACS sorted LSK. N=three individual mice/genotype (8 weeks old).