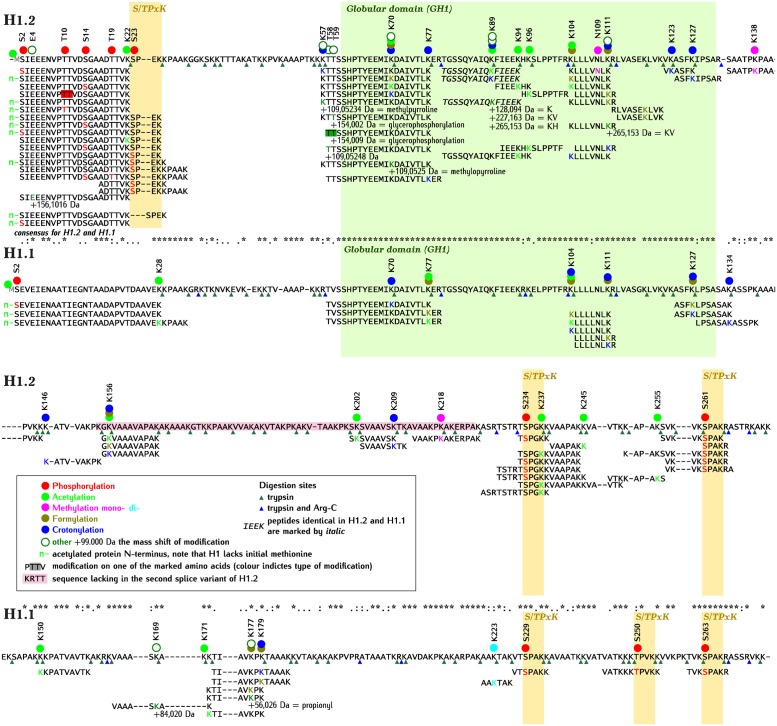
Fig 2. Alignment of amino acid sequences of Arabidopsis H1.2 and H1.1 with identified post-translational modifications.
The peptides with modifications identified by mass spectrometry are shown under the corresponding full sequence. Circles over the full sequence mark amino acids, with color filling the circle indicating the type of modification. For modifications marked “other” (white circle with dark green contour), the mass of modification in Da (eg. +99.000 Da) and the name of the possible chemical compound (eg. glycerophosphorylation) are indicated. The symbols: K, KV and KH correspond to lysine, lysine-valine and lysine-histidine, respectively. The color of letters corresponding to modified amino acids in the peptides corresponds to the type of modification, as shown for circles. Fragments of peptides identical in H1.2 and H1.1 are marked by italics. The sequences corresponding to globular domain (GH1) and S/TPxK motives are shaded in green and yellow, respectively. Sequence absent in a second splice variant of H1.2 is shaded in pink. Digestion sites by trypsin and trypsin and Arg-C proteases are marked by green and blue triangles, respectively. Green “n-” denotes acetylated protein N-terminus. Note that H1.2 and H1.1 lack initial methionine (amino acid 1) marked by grey “M” in the full sequence.