To the Editor:
Pneumonia is a widespread and severe disease and a leading cause of health care–associated infections. Staphylococcus aureus is the most important pathogen in all types of pneumonia (1). S. aureus sequence type (ST) 398 is a clone that is traditionally associated with animal farming (2). Both methicillin-sensitive (MSSA) and methicillin-resistant ST398 are predominant in livestock, but only ST398 MSSA has been reported to spread among humans (3). These potentially human-adapted ST398 MSSA strains have primarily been associated with skin and bloodstream infections (4). Notably, there are virtually no data on the role of virulence factors in isolates of the ST398 lineage achieved by advanced molecular investigation.
We performed a prospective study at the hospital of Botucatu Medical School, Brazil, from November 2011 to August 2013, that included all adult patients at the intensive care unit under mechanical ventilation (n = 270). S. aureus was isolated from 47 patients, and 27 of those patients developed pneumonia. Methicillin resistance occurred in 22 isolates (47%) and was more common in hospital-associated pneumonia (7 of 9; 78%) than in community-associated pneumonia (1 of 7; 14%) or only colonizing (without progression to respiratory infection, 6 of 18; 33%) isolates.
Multilocus sequence typing revealed that a considerable number of isolates (five; 11%) belonged to the ST398 lineage. Intriguingly, the rate of pneumonia cases was highest among the ST398 lineage (80%), as was the fatality rate resulting from pneumonia (three of four pneumonia cases; 75%). Three (27%) of 11 fatal pneumonia cases in the reported time frame were a result of ST398 isolates. All ST398 isolates were methicillin-sensitive, positive for the chp and scn genes found to be associated with human-adapted ST398 MSSA (3), and belonged to spa type t1451. Brief case reports of the three ST398-associated fatal pneumonia cases are shown in Table 1. Notably, there was at least one case of fatal ST398 pneumonia, for which the patient did not report animal contact, indicative of nosocomial acquisition of the infecting strain.
Table 1.
Brief Case Reports of Fatal Pneumonia Cases Caused by ST398 Isolates
| Patient | Isolate | Brief Case Report |
|---|---|---|
| 39-yr-old man | A16 | The patient presented at the intensive care unit for head trauma and pulmonary contusion. He developed skin and soft tissue infection, which was treated with cefepime and clindamycin. He died from pneumonia that developed within 48 h of admission. This patient was a rural worker. |
| 61-yr-old man | A19 | The patient presented at the intensive care unit for chronic renal failure with acute worsening and necessity of dialysis. He was treated with amoxicillin and clavulanic acid because of sinusitis for 3 d after admission. Afterward, the treatment was changed to imipenem and vancomycin because the patient developed ventilator-associated pneumonia. The patient died from ventilator-associated pneumonia. He reported no animal contact. |
| 76-yr-old man | A52 | The patient presented to the intensive care unit for urinary tract infection with septic shock. He received imipenem, polymyxin E, and linezolid. He died from hospital-associated pneumonia that was not associated with mechanical ventilation. He reported contact with pigs and poultry. |
Definition of abbreviation: ST = sequence type.
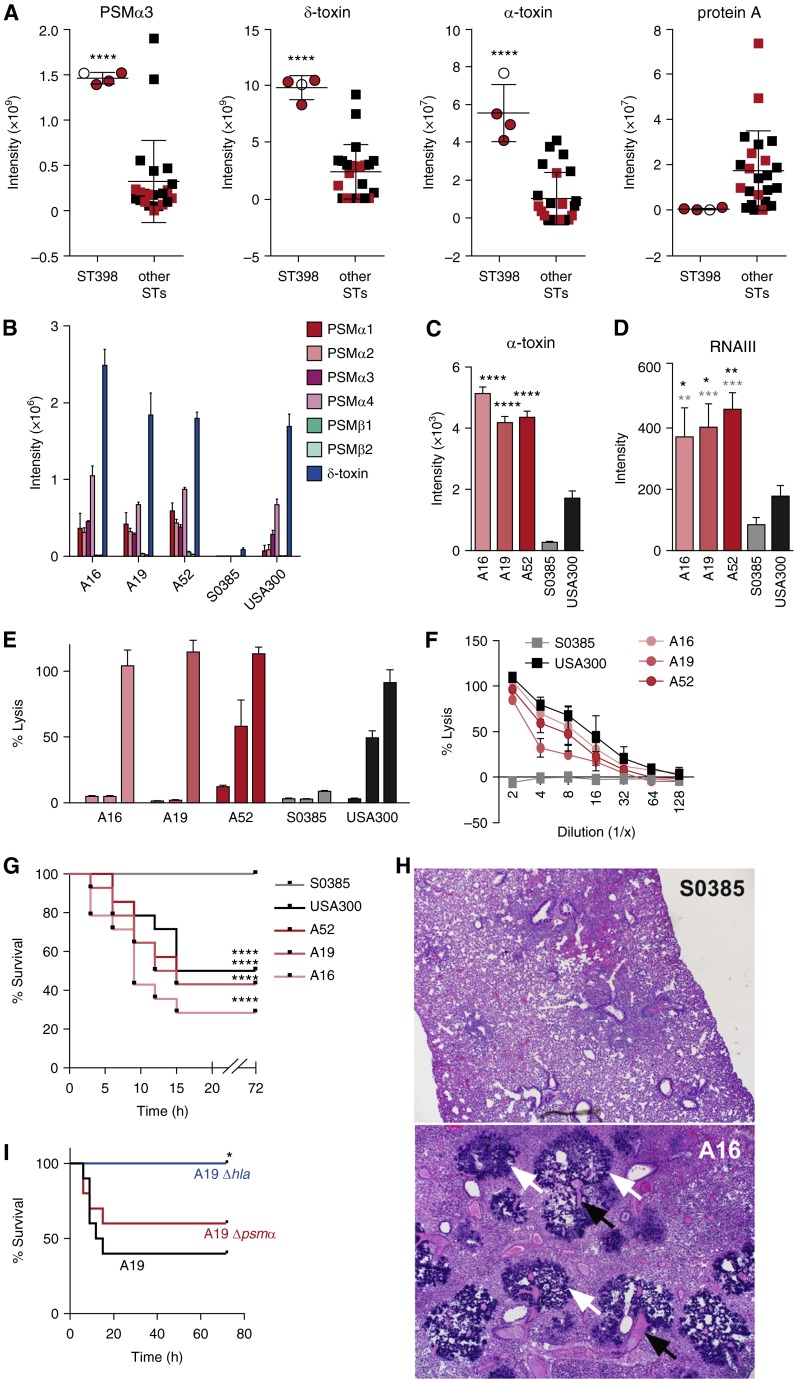
We found no significant correlation between the presence of key virulence genes and pneumonia or fatality. Prompted by our recent findings that the severity of staphylococcal skin infection is influenced to a considerable degree by gene expression levels (5), we hypothesized that this may also be the case for ST398 pneumonia. We focused on the major factors shown to affect staphylococcal lung infection: α-toxin, phenol-soluble modulin (PSM) peptides, and protein A (6–8). The genes encoding Panton-Valentine leukocidin were not present in our ST398 isolates. When we compared all pneumonia isolates, the ST398 isolates showed significantly higher in vitro expression levels of the most cytolytic PSM, PSMα3, δ-toxin (a readout for functionality of the accessory gene regulator Agr), and α-toxin, than non-ST398 isolates (Figure 1A). Differences in other PSMs were similar (data not shown). Furthermore, PSM and α-toxin levels were similar to or exceeded those detected in the highly virulent community-associated methicillin-resistant S. aureus clone USA300, which is known to strongly express PSMs and α-toxin and whose virulence potential relies on those cytolysins (6, 9) (Figures 1B and 1C). Notably, ST398 PSM and α-toxin levels strongly exceeded those detected in the standard ST398 strain S0385, a Dutch livestock-associated methicillin-resistant S. aureus isolate from a case of human endocarditis (10). In contrast, production of the negatively Agr-regulated protein A was significantly lower in the ST398 isolates compared with the other isolates (Figure 1A). Thus, our data were indicative of a highly functional Agr system in the ST398 isolates, which we confirmed by quantitative real-time polymerase chain reaction of RNAIII (Figure 1D). Furthermore, among isolates that caused fatal pneumonia, high production of PSMα and α-toxin only occurred in ST398 strains, indicating there is a specific role for PSMα peptides and α-toxin in the extraordinary virulence of ST398 isolates. The ST398 isolates showed pronounced capacities to lyse human erythrocytes and neutrophils (Figures 1E and 1F), demonstrating that high cytolysin expression levels translate to strong cytolytic capacities.
Figure 1.
Molecular investigation of fatal pneumonia resulting from sequence type (ST) 398. (A) In vitro production levels of phenol-soluble modulin (PSM) α3 and δ-toxin (by high-performance liquid chromatography/mass spectrometry) and α-toxin and protein A (by Western blot densitometry) of all pneumonia isolates (here and all other experiments: grown to stationary growth phase in tryptic soy broth). Values corresponding to fatal cases are shown in red. ****P < 0.0001 (unpaired t test). (B and C) Comparison of in vitro production levels of PSMs (B) and α-toxin (C) to those of USA300 (the USA300 isolate SF8300 was used for all analyses in the present study) and the ST398 endocarditis isolate S0385. ****P < 0.0001 (vs. USA300 and S0385, one-way analysis of variance [ANOVA]). (D) RNAIII expression levels by quantitative real-time polymerase chain reaction. *P < 0.05; **P < 0.01; ***P < 0.001 (one-way ANOVA; black asterisks, vs. USA300; gray asterisks, vs. S0385). (E) Neutrophil lysis. Culture filtrates were measured for their capacity to lyse human neutrophils by release of lactate dehydrogenase. The three bars for each strain represent decreasing dilutions: 1:10; 1:5; and 1:2. (F) Lysis of human erythrocytes. Culture filtrates were used at the given dilutions. (G–I) Mouse pneumonia model. Mice were infected intranasally with 1 × 108 colony-forming units, and disease development was monitored for a total of 72 hours. (G) Comparison of ST398 pneumonia isolates with USA300 and S0385. n = 25 mice per group. ****P < 0.0001 (log-rank test, vs. data obtained with S0385). (H) Histology. Microscopic images of lung tissue; magnification 40×. White arrows point to bacterial colonies, black arrows to necrotic vessels. An image of lung tissue of a mouse infected with strain A16 is shown; pathology in the lungs of mice infected with other ST398 strains and USA300 were similar. (I) Comparison of ST398 A19 isolate wild-type versus isogenic hla and psmα deletion strains. n = 10 mice per group. *P < 0.05 (log-rank test; vs. data obtained with wild-type strain A19). (A–F) Error bars show ±SD.
In a mouse pneumonia model (Figure 1G), the fatality rate in mice infected with the three ST398 strains was high, at least equaling that caused by strain USA300, and significantly higher than that caused by strain S0385. Accordingly, histological analysis demonstrated extensive pathology in the lungs of mice infected with the ST398 or USA300 strains, which all showed signs of severe and multifocal necrotizing pneumonia, with acute inflammation, necrotizing vasculitis, thrombosis, and large numbers of bacterial colonies, to an extent that was indistinguishable between groups (shown in Figure 1H for strain A16). Mice infected with strain S0385, in contrast, showed only signs of moderate peribronchiolar pneumonia and no bacterial colonies (Figure 1H). Thus, the murine pneumonia model well reflected the clinical results and confirmed the high virulence of the ST398 isolates.
Then, to analyze the effect of PSMα and α-toxin, we produced isogenic gene deletion mutants in the psmα operon and hla gene in isolate A19. Both psmα and hla mutants caused a lower number of deaths than the wild-type A19 isolate (Figure 1I). However, only the hla mutant survival rates were significantly different from those of the wild-type strain, with no animals infected by the A19 hla mutant succumbing to the infection. These results attribute a central function to α-toxin and high α-toxin expression in the pathogenesis of fatal pneumonia by ST398 S. aureus.
In conclusion, our study identifies ST398 MSSA as a dangerous and highly virulent emerging source of fatal pneumonia. Furthermore, although our results emphasize strain dependence, they are in accordance with the notion of a generally crucial role of α-toxin in lung infection. Finally, our findings call for surveillance measures analyzing not only antibiotic resistance but also virulence potential of S. aureus as an important factor that contributes to infection outcome.
Acknowledgments
Acknowledgment
The authors thank A. C. Fluit, Utrecht Medical Center, for providing strain S0385; Binh Diep, University of California, San Francisco, for isolate SF8300; and Dan Long and Dana P. Scott, Pathology Branch, Rocky Mountain Laboratories, National Institute of Allergy and Infectious Diseases, for histological analyses.
Footnotes
M.F.B. received funding from the CAPES Ph.D. program of the Brazilian government (grant 99999.007555/2013-00). M.O. received funding from the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (grant ZIA AI000904-14).
Author Contributions: M.F.B., A.J.Y., A.E.V., J.M., H.-S.J., and G.Y.C.C. performed experiments; M.T.S. and C.F.R. obtained patient samples; C.M.C.B.F. and R.S.C. collected clinical data; M.F.B., H.-S.J., G.Y.C.C., and M.O. analyzed data; M.F.B., M.L.R.S.C., and M.O. conceived the study and supervised experiments; and M.O. wrote the paper.
Author disclosures are available with the text of this letter at www.atsjournals.org.
References
- 1.Kollef MH, Shorr A, Tabak YP, Gupta V, Liu LZ, Johannes RS. Epidemiology and outcomes of health-care-associated pneumonia: results from a large US database of culture-positive pneumonia. Chest. 2005;128:3854–3862. doi: 10.1378/chest.128.6.3854. [DOI] [PubMed] [Google Scholar]
- 2.Smith TC, Pearson N. The emergence of Staphylococcus aureus ST398. Vector Borne Zoonotic Dis. 2011;11:327–339. doi: 10.1089/vbz.2010.0072. [DOI] [PubMed] [Google Scholar]
- 3.Uhlemann AC, Porcella SF, Trivedi S, Sullivan SB, Hafer C, Kennedy AD, Barbian KD, McCarthy AJ, Street C, Hirschberg DL, et al. Identification of a highly transmissible animal-independent Staphylococcus aureus ST398 clone with distinct genomic and cell adhesion properties. MBio. 2012;3:3. doi: 10.1128/mBio.00027-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uhlemann AC, Hafer C, Miko BA, Sowash MG, Sullivan SB, Shu Q, Lowy FD. Emergence of sequence type 398 as a community- and healthcare-associated methicillin-susceptible Staphylococcus aureus in northern Manhattan. Clin Infect Dis. 2013;57:700–703. doi: 10.1093/cid/cit375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li M, Diep BA, Villaruz AE, Braughton KR, Jiang X, DeLeo FR, Chambers HF, Lu Y, Otto M. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci USA. 2009;106:5883–5888. doi: 10.1073/pnas.0900743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bubeck Wardenburg J, Bae T, Otto M, Deleo FR, Schneewind O. Poring over pores: alpha-hemolysin and Panton-Valentine leukocidin in Staphylococcus aureus pneumonia. Nat Med. 2007;13:1405–1406. doi: 10.1038/nm1207-1405. [DOI] [PubMed] [Google Scholar]
- 7.Diep B.Relative effects of Panton Valentine leukocidin, alpha-hemolysin and alpha-type phenol-soluble modulins in pathogenesis of community-associated MRSA necrotizing pneumonia. Presented at the International Conference on Antimicrobial Agents and Chemotherapy. September 12–15, 2010, Boston, MA [Google Scholar]
- 8.Gómez MI, O’Seaghdha M, Magargee M, Foster TJ, Prince AS. Staphylococcus aureus protein A activates TNFR1 signaling through conserved IgG binding domains. J Biol Chem. 2006;281:20190–20196. doi: 10.1074/jbc.M601956200. [DOI] [PubMed] [Google Scholar]
- 9.Wang R, Braughton KR, Kretschmer D, Bach TH, Queck SY, Li M, Kennedy AD, Dorward DW, Klebanoff SJ, Peschel A, et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13:1510–1514. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- 10.Schijffelen MJ, Boel CH, van Strijp JA, Fluit AC. Whole genome analysis of a livestock-associated methicillin-resistant Staphylococcus aureus ST398 isolate from a case of human endocarditis. BMC Genomics. 2010;11:376. doi: 10.1186/1471-2164-11-376. [DOI] [PMC free article] [PubMed] [Google Scholar]