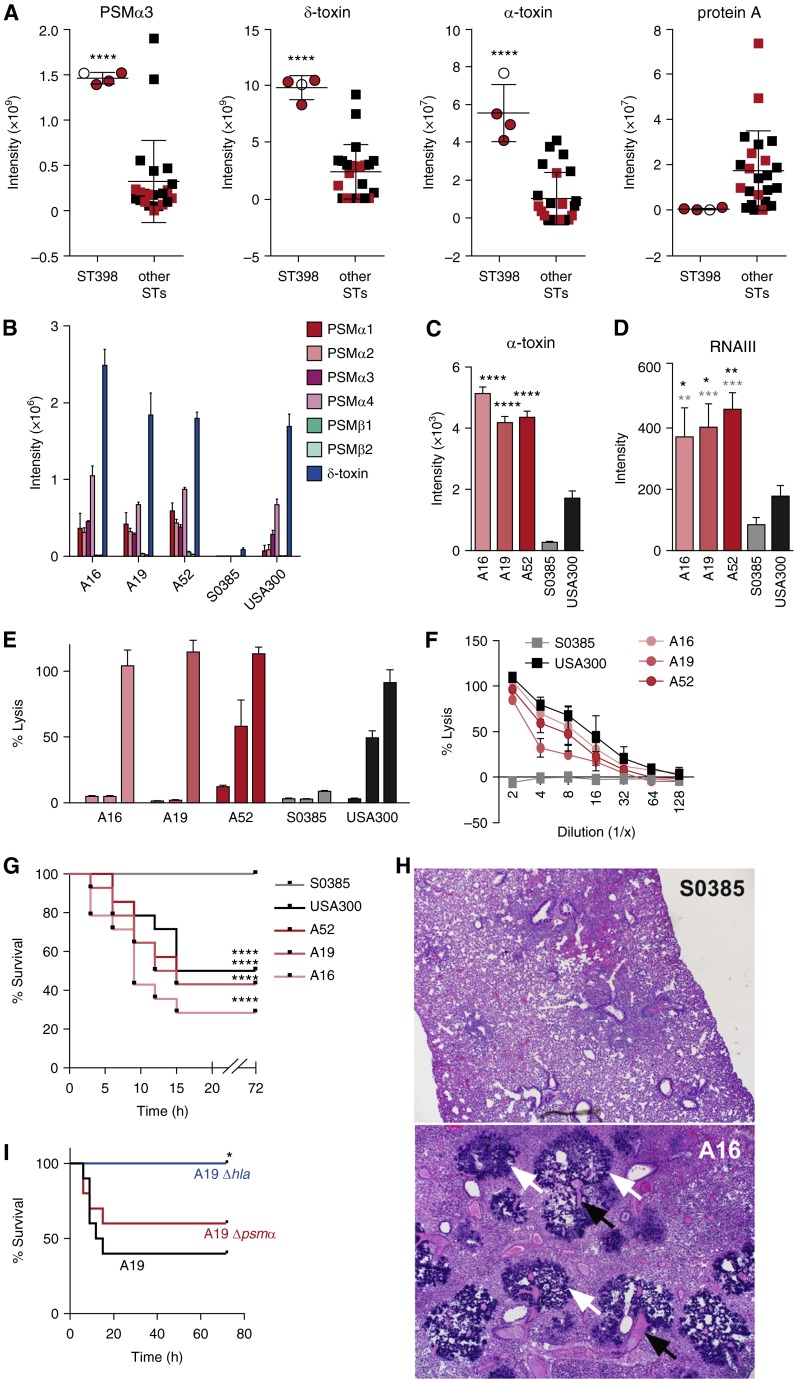
Figure 1.
Molecular investigation of fatal pneumonia resulting from sequence type (ST) 398. (A) In vitro production levels of phenol-soluble modulin (PSM) α3 and δ-toxin (by high-performance liquid chromatography/mass spectrometry) and α-toxin and protein A (by Western blot densitometry) of all pneumonia isolates (here and all other experiments: grown to stationary growth phase in tryptic soy broth). Values corresponding to fatal cases are shown in red. ****P < 0.0001 (unpaired t test). (B and C) Comparison of in vitro production levels of PSMs (B) and α-toxin (C) to those of USA300 (the USA300 isolate SF8300 was used for all analyses in the present study) and the ST398 endocarditis isolate S0385. ****P < 0.0001 (vs. USA300 and S0385, one-way analysis of variance [ANOVA]). (D) RNAIII expression levels by quantitative real-time polymerase chain reaction. *P < 0.05; **P < 0.01; ***P < 0.001 (one-way ANOVA; black asterisks, vs. USA300; gray asterisks, vs. S0385). (E) Neutrophil lysis. Culture filtrates were measured for their capacity to lyse human neutrophils by release of lactate dehydrogenase. The three bars for each strain represent decreasing dilutions: 1:10; 1:5; and 1:2. (F) Lysis of human erythrocytes. Culture filtrates were used at the given dilutions. (G–I) Mouse pneumonia model. Mice were infected intranasally with 1 × 108 colony-forming units, and disease development was monitored for a total of 72 hours. (G) Comparison of ST398 pneumonia isolates with USA300 and S0385. n = 25 mice per group. ****P < 0.0001 (log-rank test, vs. data obtained with S0385). (H) Histology. Microscopic images of lung tissue; magnification 40×. White arrows point to bacterial colonies, black arrows to necrotic vessels. An image of lung tissue of a mouse infected with strain A16 is shown; pathology in the lungs of mice infected with other ST398 strains and USA300 were similar. (I) Comparison of ST398 A19 isolate wild-type versus isogenic hla and psmα deletion strains. n = 10 mice per group. *P < 0.05 (log-rank test; vs. data obtained with wild-type strain A19). (A–F) Error bars show ±SD.