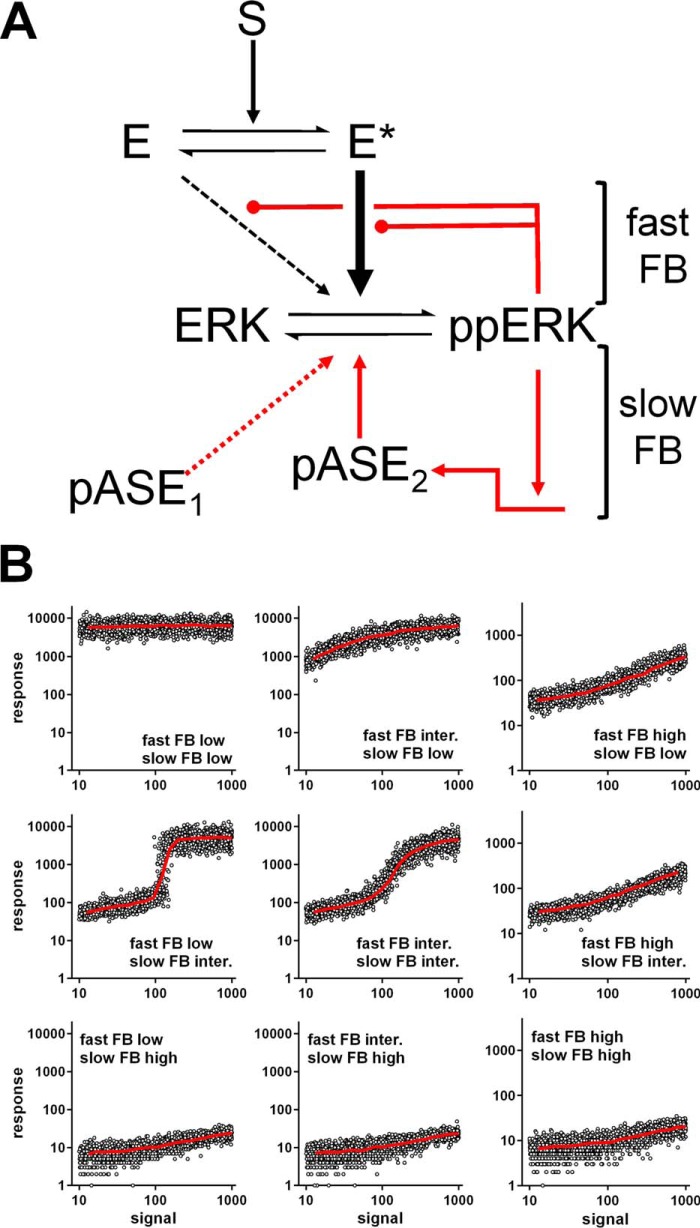
FIGURE 3.
Stochastic modeling of signaling to ERK. Panel A, ERK signaling model incorporating two negative feedback loops: a fast one from ppERK to effector E and a slow one via phosphatase (p-ASE2) expression. Panel B, we ran stochastic simulations of this model using the parameters in Table 1, varying the strength of the fast and slow negative feedback (FB) loops and for each condition using 2000 independent runs of the model. For each run we sampled the total ERK levels from a log10 normal distribution (mean of 3.9 and a S.D. 0.1) reaching steady state before signal introduction. Signal molecule number was randomly drawn from a uniform distribution on the log10 scale between 10 and 1000 molecules, and for all simulations the stochastic simulation (Gillespie) algorithm was used, as described under “Experimental Procedures.” The figure shows data for 60 min stimulations with fast FB set at low, intermediate, or high strength (0.01, 1.66, and 330, respectively), and slow FB was set at low, intermediate, and high strength (1, 100, and 10000, respectively) with the line of best fit (red) superimposed over individual run values (open circles). These are from a larger series of simulations run for 5–360 min with a wider range of fast FB and slow FB values, from which the MI values shown in Fig. 4 and Fig. 12 were calculated.