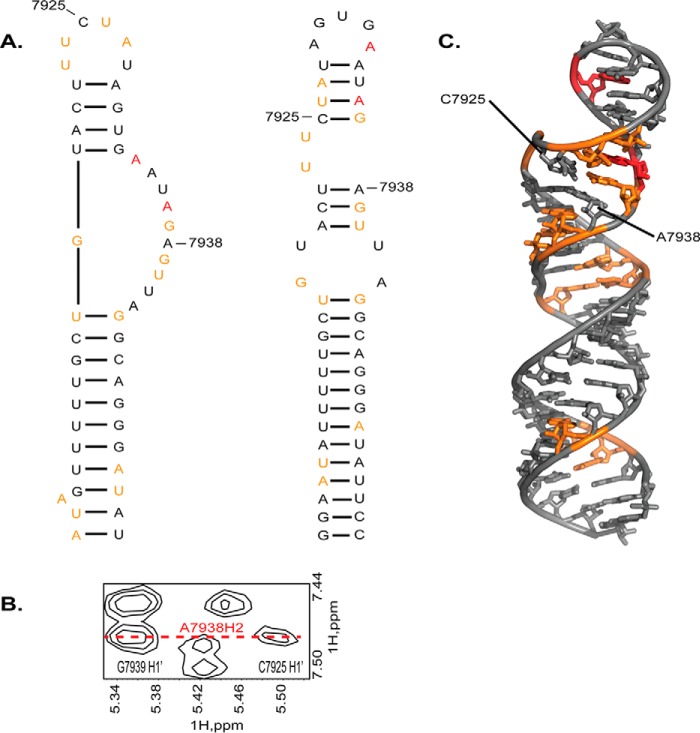
FIGURE 10.
Comparison of SHAPE-calculated and phylogenetic models of SL1ISS. A, left, secondary structure of SL1ISS determined by SHAPE modification and covariation analysis within the context of the entire HIV-1 genome (17). Right, alternative model of SL1ISS determined by phylogenetic comparisons and chemoenzymatic probing of the isolated ssA7 locus (12, 13). B, NOE evidence that the upper helix of SL1ISS folds with a structure consistent with the phylogenetic model. C, mapping of the SHAPE reactivity data onto the three-dimensional structure of SL1ISS shows that ∼87% of the low and medium reactive nucleotides can be accommodated in regions of stable structure.