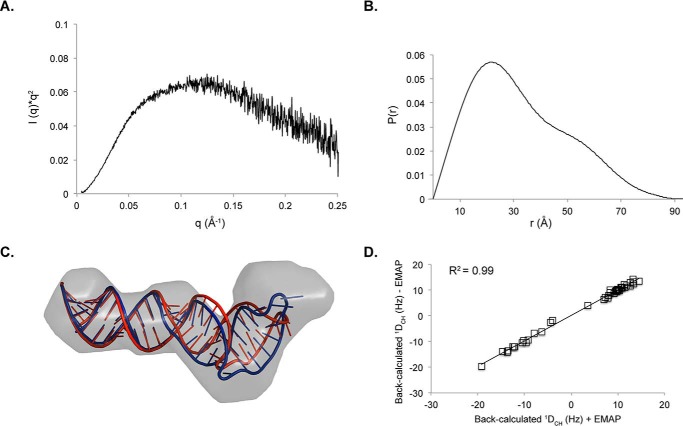
FIGURE 7.
Small angle x-ray scattering of SL1ISS. A, Kratky profile of SL1ISS demonstrates that the RNA adopts a stable fold in solution. B, pair distance distribution function of SL1ISS. C, refinement of SL1ISS NMR structure against the SAXS reconstruction using AMBER. The SAXS molecular envelope is shown as a transparent surface with the lowest energy SL1ISS NMR structure (red) fit to the density. The NMR structure was jointly refined against NOE distance restraints, RDCs, and the molecular reconstruction (blue) using the EMAP force field term in AMBER. The two structures superimpose over all base pairs from the lower helix with an r.m.s.d. of 1.3 Å. D, correlation plot between back-calculated RDC values with (x axis) and without (y axis) inclusion of the molecular reconstruction during refinement.