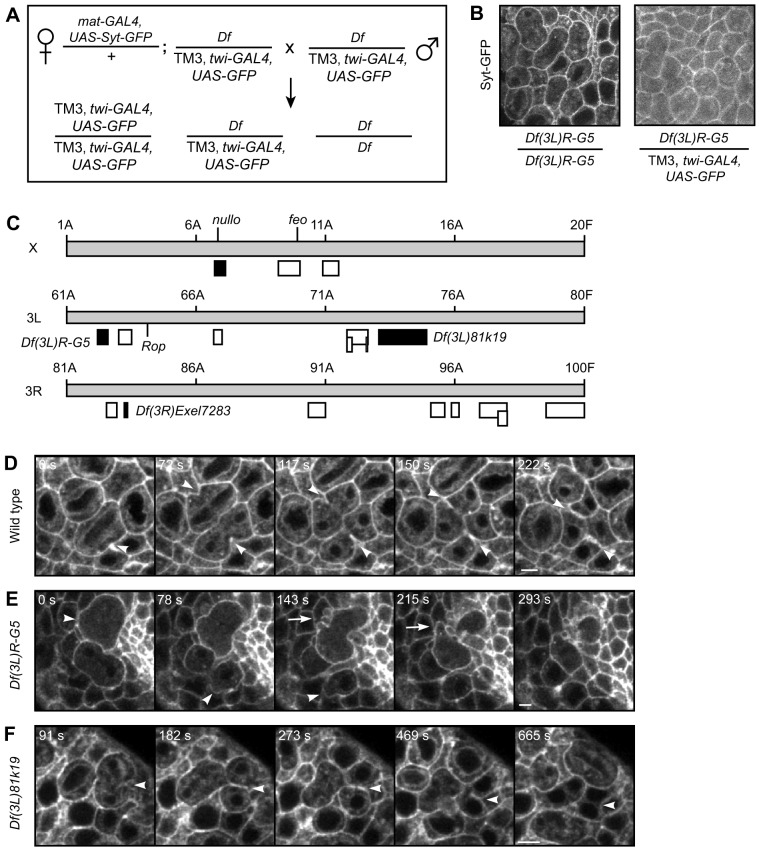
Fig. 2.
Cytokinesis defects identified in a live-imaging screen of Drosophila embryos. (A) Genotypes and crossing scheme used to create homozygous deficiency embryos maternally loaded with Synaptotagmin–GFP (Syt–GFP). mat-GAL4 drives maternal loading, twist(twi)-GAL4 drives embryonic expression. (B) GFP signal of mitotic domains in cycle 14 embryos either homozygous or heterozygous for Df(3L)R-G5. Homozygous embryos were identified by the absence of ubiquitous cytoplasmic GFP from the twist-GAL4, UAS-GFP balancer chromosome. Maternally loaded Syt–GFP marks cell membranes in both embryos. (C) Diagram of the X and third chromosomes, shown as gray boxes, with cytological numbers indicated above. Boxes below the chromosomes depict the deficiency lines screened. Black indicates phenotype observed. White indicates no phenotype. Location of the three genes nullo, feo and Rop are also noted. (D–F) Time-lapse images of Syt–GFP in cycle 14 mitotic domains of wild-type (D, Movie 1), Df(3R)R-G5 (E, Movie 2) and Df(3L)81k19 (F) embryos. Syt–GFP marks the plasma membrane and ingressing furrow. The time from the beginning of furrow initiation to the first cell dividing is indicated. (D) Normal furrow ingression (arrowheads) in wild-type cells. (E) In Df(3L)R-G5 embryos, cells undergoing cytokinesis displayed abnormally large membrane blebs (arrowheads) and ectopic blebs at the furrow (arrow). (F) In Df(3L)81k19 embryos, cytokinesis furrows regressed (arrowheads). Scale bars: 5 µm.