Abstract
We recently reported on our success to generate deletion mutants of the genes encoding glutamate dehydrogenase (GDH) and glutamine oxoglutarate aminotransferase (GOGAT) in M. bovis BCG, despite their in vitro essentiality in M. tuberculosis. We could use these mutants to delineate the roles of GDH and GOGAT in mycobacterial nitrogen metabolism by using M. bovis BCG as a model for M. tuberculosis specifically. Here, we extended our investigation towards the involvement of GDH and GOGAT in other aspects of M. bovis BCG physiology, including the use of glutamate as a carbon source and resistance to known phagosomal stresses, as well as in survival inside macrophages. We find that gdh is indispensable for the utilization of glutamate as a major carbon source, in low pH environments and when challenged with nitric oxide. On the other hand, the gltBD mutant had increased viability under low pH conditions and was unaffected by a challenge with nitric oxide. Strikingly, GDH was required to sustain M. bovis BCG during infection of both murine RAW 264.7 and bone-marrow derived and macrophages, while GOGAT was not. We conclude that the catabolism of glutamate in slow growing mycobacteria may be a crucial function during infection of macrophage cells and demonstrate a novel requirement for M. bovis BCG GDH in the protection against acidic and nitrosative stress. These results provide strong clues on the role of GDH in intracellular survival of M. tuberculosis, in which the essentiality of the gdh gene complicates knock out studies making the study of the role of this enzyme in pathogenesis difficult.
Introduction
Rapidly increasing numbers globally of tuberculosis (TB) patients infected with Mycobacterium tuberculosis strains resistant to many TB drugs underscores an urgent need for the development of new TB chemotherapies acting by mechanisms that are distinct from the current anti-TB drugs [1]. The central carbon and nitrogen metabolism of M. tuberculosis is composed of unique pathways that often are crucial to the survival of this organism and therefore represents a reservoir of enzymes that may be exploited as targets for novel anti-TB chemotherapy development [2,3].
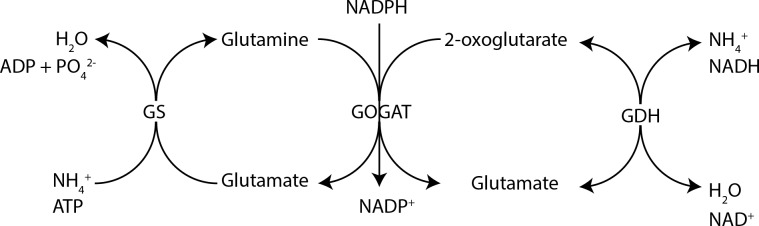
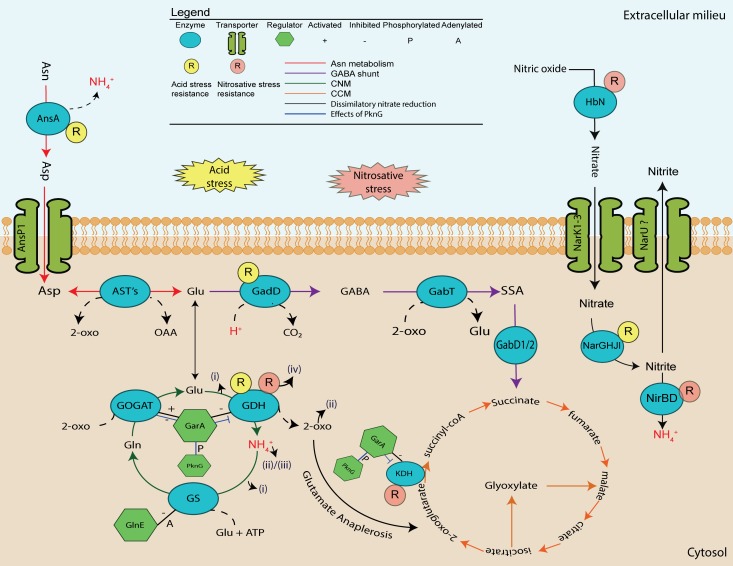
In particular the involvement of protein kinase G (PknG) and glycogen accumulation factor A (GarA), has been implicated in the regulation of enzymes that are effectors of central carbon metabolism (CCM) and central nitrogen metabolism (CNM) [4–8]. It was found that binding of GarA to the ketoglutarate dehydrogenase complex (KDH), glutamate dehydrogenase (GDH) and glutamine oxoglutarate aminotransferase (GOGAT), inhibits KDH and GDH activity, while it stimulated GOGAT activity [6,7]. Activated GOGAT promotes glutamate accumulation and subsequently removes 2-oxoglutarate which results in cataplerosis of the TCA cycle and nett glutamate accumulation (Fig 1) [9]. Phosphorylation of GarA by PknG abrogated the interaction between GarA and KDH/GDH/GOGAT resulting in nett glutamate degradation and accumulation of 2-oxoglutarate resulting in anaplerosis of TCA cycle intermediates [6,7,9]. These results implicated regulation of glutamate metabolism as an important factor of M. tuberculosis physiology. Strikingly the genes encoding GDH and GOGAT were found to be essential for in vitro growth of M. tuberculosis further emphasising a requirement for homeostatic regulation of glutamate by this pathogen [10–12]. However, a mutant of the gene encoding PknG could be obtained in M. tuberculosis allowing the delineation of the role of this virulence factor in pathogenesis [4]. An M. smegmatis conditional garA mutant was also recently generated to study the role of GarA in mycobacterial physiology more clearly, but its essentiality in M. tuberculosis has complicated efforts to study its involvement in pathogenesis [8]. Similarly the essentiality of gdh and gltBD has probably hindered attempts to more clearly define their specific contributions to pathogenicity. However, we recently reported on our success to generate mutants of these genes in M. bovis BCG, which is highly similar to M. tuberculosis at a genetic level [13]. With the use of these mutants we could delineate the roles of these two enzymes in the nitrogen metabolism of M. bovis BCG, by showing that GOGAT was essential for growth on ammonium as a sole nitrogen source, while GDH was required for optimal growth on glutamate as a sole nitrogen source and when high levels of asparagine was present in the growth medium [13]
Fig 1. Schematic illustration depicting central nitrogen metabolism in the mycobacterium tuberculosis complex.
Here we have used our M. bovis BCG gdh and gltBD mutants to investigate more clearly their importance in intracellular survival by using in vitro conditions that simulate some of the stresses employed by macrophages to kill infecting mycobacteria. We demonstrate an absolute requirement for gdh under limiting carbon conditions when glutamate was a major carbon source. We also demonstrate a protective role for gdh under nitrosative and acidic stress conditions. Surprisingly, gltBD was dispensable for survival under all of the aforementioned conditions. Finally, we use two murine macrophage systems to demonstrate that gdh is required for optimal survival of M. bovis BCG inside macrophages. Ours is the first report to implicate gdh in the intracellular survival of a mycobacterium.
Our results indicate that the catabolism of glutamate is an important process in M. bovis BCG which is primarily catalysed by the gdh protein product. While the enzyme product of the gltBD operon, glutamine oxoglutarate aminotransferase (GOGAT) has received some attention as a possible anti-TB drug target due to inhibition of M. tuberculosis by azaserine (an inhibitor of GOGAT) in vitro [14,15], our findings suggest that GDH may be as important if not more to the survival of M. tuberculosis inside host macrophage cells. Notably, the M. tuberculosis GDH is structurally and functionally different from the GDH found in humans as well as in the human intestinal flora, which may make this enzyme a potential specific target for anti-TB drug intervention [15].
Materials and Methods
Cultivation of bacteria
All bacterial strains used are listed in S2 Table. M. bovis BCG was cultured without agitation in 7H9 (BD scientific, USA) supplemented with 0.05% Tween 80, 0.2% glycerol, 5 g/L albumin, 2 g/L glucose and 1.5 mg/L catalase in 25 cm2 cell culture flasks (Nunc, Denmark). The ΔgltBD mutant which had a growth deficiency in standard 7H9 was maintained in 7H9 supplemented with 20 mM L-glutamate. In certain cases (indicated in the text), bacterial strains were cultivated in 7H9 that was not supplemented with Tween 80, glycerol, albumin, glucose or catalase. Genetically complemented strains were maintained under selection with gentamycin at 2.5 μg/ml.
Nitric oxide challenge
All strains were challenged with diethelenetriamine/nitric oxide adduct (DETE/NO) (Sigma-Aldrich, USA). DETE/NO was stored as at -20°C as a lyophilised powder prior to use. The nitric oxide donor was prepared by solubilising DETE/NO in ultra-pure water to a final concentration of 5 M and immediately used as required.
M. bovis BCG, Δgdh, Δgdh: complement, ΔgltBD and ΔgltBD complement were cultivated as explained in the cultivation of bacteria section albeit slightly modified. OADC was replaced with ADN (Bovine serum albumin fraction V, 50g/L; D-dextrose, 20 g/L; sodium chloride, 8.5 g/L) as an alternative enrichment to avoid catalase. Upon reaching an OD600 of 0.8–1.0 all cultures were diluted to an OD600 of 0.0005 in 5 ml 7H9 supplemented with ADN and tween followed by challenge with 500 μM DETE/NO for a total period of 48 hours. Cells were plated on Middlebrook 7H11 agar (BD scientific, USA) supplemented with 0.2% glycerol and OADC at various time points (0h, 6h, 12h, 24h, 36h, 48h) and cell viability was determined by CFU.
Culturing and infection of RAW 264.7 macrophages
RAW 264.7 ATCC® TIC-71™ macrophages (ATCC, USA) were maintained in DMEM (Lonza, USA) at 37°C under 5% CO2 until confluent. Viable RAW 264.7 ATCC TIB-71 cells were quantified by staining with trypan blue and counted using a haemocytometer. Macrophages in DMEM supplemented with 10% FBS and 10 μg/ml LPS (Sigma Aldrich, USA) (D10) were seeded at 5 x 105 cells/well in 24 well plates (Nunc, Germany) and incubated at 37°C under 5% CO2 overnight to allow the cells to adhere. Bacteria cultured to an OD600 of 0.8–1.0 in standard 7H9 without agitation at 37°C were pelleted and re-suspended in equal volumes of D10, which was then adjusted to an OD600 of 1.0 (corresponding to approximately 2.5 × 107 CFU/100 μl). Bacteria were added directly to RAW 264.7 macrophage monolayers to obtain a multiplicity of infection (MOI) of 5:1 bacteria to macrophage and incubated at 37°C under 5% CO2 for 3 hours. The culture medium was subsequently removed and replaced with fresh D10 medium supplemented with penicillin/streptomycin (penstrep) antibiotic cocktail (1:100) and incubated for an additional hour at 37°C under 5% CO2. Monolayers were subsequently washed with PBS and supplemented with fresh D10. Infected RAW 264.7 macrophages were lysed by the addition of ultra-pure sterile water and scraping. Lysates were serially diluted prior to plating on 7H11 agar (BD scientific, USA). Enumerations of CFU were performed after approximately 2 weeks of culturing at 37°C when colonies started to appear.
Culturing and infection of murine bone-marrow derived macrophages (BMDM)
Culturing and infection of BMDM macrophages was performed as previously described with some modifications [16]. Tibia and femur bones from 6–10 week old female C57/BL6 mice bred and housed under specific pathogen-free conditions were obtained from the Stellenbosch University animal facility. The extraction of bones from the aforementioned mice was carried out in strict accordance with the recommendations as reflected in the South African National Standards 10386: 2008.
The protocol, SU-ACUD14-00041, was approved by the research ethics committee: animal care and use of the University of Stellenbosch. Mice were euthanized prior to the extraction of tibia and femur bones by cervical dislocation. The bones were dissected and the marrow flushed out and dispersed with 2.5 ml of RPMI 1640 medium with L-glutamine and sodium bicarbonate (Sigma, USA) and a 25-GA syringe needle. The bone marrow cell suspension was adjusted to 2 × 105 cells/ml in RPMI-LCSF20 medium (consisted of RPMI 1640 medium with L glutamine and sodium bicarbonate, 10% fetal bovine serum (Biochrom, UK) and 20% supernatant from L929 cells. The cells were plated in 10 ml volumes on 90 mm diameter Petri dishes (Greiner, USA) and incubated at 37°C with 5% CO2 for 7 days. Cells were fed with 10 ml RPMI-LCSF after 4 days. Non-adherent cells were removed in two washes with PBS (Lonza, USA) before adherent cells were released with 10 ml ice cold PBS. Released macrophages were adjusted to 2 × 105 cells/ml in RPMI-LCSF10 (as described before but with 10% supernatant from L929 cells). Cells were seeded in 24 well plates (1 ml/well) and allowed to adhere for 24 hours. Bacteria grown to early exponential growth phase (OD600 = 0.5–0.8) were washed once with PBS (Lonza, USA). Bacterial aggregates were removed by low speed centrifugation (200 × g for 10 minutes) and the resulting supernatant passed through a 5 μm surfactant free cellulose acetate syringe filter (Sartorius, Germany). Bacterial suspensions were prepared in RPMI-LCSF10 to yield an MOI of 1:1. At this point the macrophage culture medium was replaced with 0.5 ml bacterial suspension. Infection of macrophages lasted for 4h before they were washed four times with pre-warmed PBS to eliminate non-phagocytosed bacteria and re-fed with RPMI-LCSF10. The medium covering each monolayer was replaced every 48h. Infected macrophages were lysed with 0.1% Triton X-100 solution (Sigma, Germany). Serial dilutions of the lysates were prepared in 7H9 and spread on 7H11 agar supplemented with OADC (BD scientific, USA). Bacterial colonies became visible and were enumerated after approximately 2 weeks of incubation at 37°C.
Statistical analysis
Statistical analyses were carried out with the statistics software GraphPad Prism 5 version 5.01 (GraphPad Software). All statistical tests performed are indicated in figure legends. Probabilities of < 0.05 were considered significant.
Results
gdh is required for the utilization of glutamate as a major carbon source
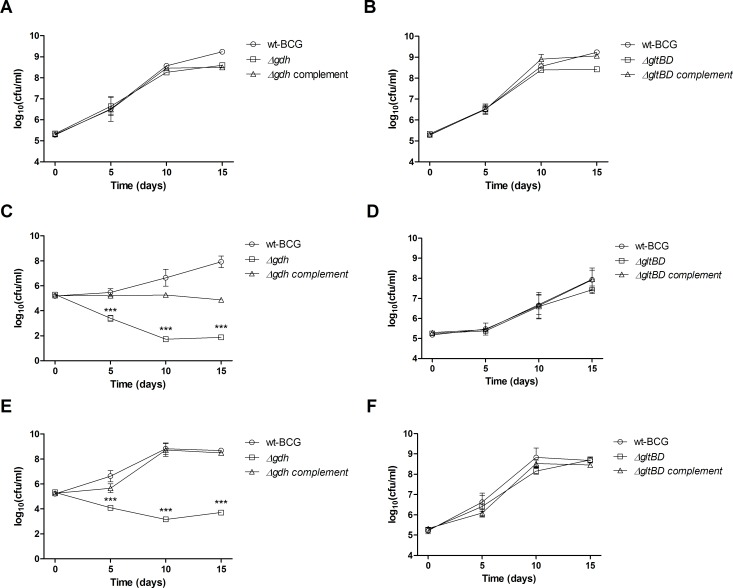
We previously reported GDH’s importance for optimal growth of M. bovis BCG when glutamate was the sole nitrogen source and when cultured in the presence of excessive asparagine levels [13]. Since there is some evidence that glutamate may also be utilized as a carbon source by M. tuberculosis [17,18], we analysed the growth of M. bovis BCG in 7H9 medium which was not supplemented with albumin, dextrose or glycerol and in which the detergent was tyloxapol as opposed to tween 80. This medium, which contains glutamate as the major carbon source at 3.4 mM in addition to the carbon sources citrate (0.5 mM), pyridoxine (2.9 μM) and biotin (4.1 μM), could support growth of M. bovis BCG (Fig 2C), although the replication rate was lower than when the medium was supplemented with albumin, dextrose, glycerol and Tween 80 (Fig 2A). Strikingly the Δgdh mutant became non-viable in the medium containing glutamate as major carbon source with an approximately 3 log10 (CFU/ml) reduction in viable cell counts after 10 days of culture (Fig 2C). Notably this phenotype of the Δgdh mutant is much more severe than the phenotype we previously reported for the same mutant when it was cultured in medium containing glutamate as the sole nitrogen source, in which case replication rate was merely decreased and cultures reached a lower maximum density compared to wild type [13]. Also in contrast to earlier observations in medium containing glutamate as sole nitrogen source, the decrease in viability of the Δgdh mutant in medium containing glutamate as major carbon source was not completely reversed by re-introduction of an intact copy of gdh as the complemented strain remained static over the course of the experiment. This poor complementation corresponded with low specific GDH activity levels in crude lysates as well as lower mRNA expression of gdh in the genetically complemented strain compared to the wild type strain (S1 Table). It was shown that M. tuberculosis uses cholesterol as a major carbon source during infection of mice [19]. Therefore, we tested whether the addition of cholesterol to culture medium containing glutamate as major carbon source would chemically complement for the loss in viability suffered by the Δgdh mutant. Unexpectedly the Δgdh mutant continued to become non-viable in the medium containing cholesterol in addition to the major carbon source glutamate (Fig 2E), while the ΔgltBD mutant exhibited no apparent growth defects under the conditions tested (Fig 2B, 2D & 2E). However, addition of cholesterol improved growth of the Δgdh complemented strain to wild type levels (Fig 2E). Collectively these results indicate that GDH is indispensable for growth of M. bovis BCG under conditions when nutrients are limiting and some of the only available sources of both nitrogen and carbon is glutamate and related amino acids, such as glutamine; asparagine and aspartate that are easily converted to glutamate.
Fig 2. The effect of glutamate as a major carbon source on survival of M. bovis BCG wild type, mutant (Δgdh and ΔgltBD) and complemented strains.
Exponential phase cultures were diluted to an OD600 of 0.0005 into (A & B) 7H9 supplemented with glycerol, dextrose and Tween 80, (C & D) 7H9 without glycerol, dextrose, or Tween 80 (replaced with tyloxapol) or (E & F) 7H9 without glycerol, dextrose, or Tween80 and supplemented with cholesterol (0.25 mM). Symbols and error bars are means and standard errors calculated from triplicate plating obtained from two independent experiments. Data was analysed by a regular two-way ANOVA test with Bonferroni post-testing to compare mean CFU/ml of mutant (Δgdh and ΔgltBD) and wild type strains. * p < 0.05, ** p < 0.01, *** p < 0.001.
Inactivation of gdh increases sensitivity to low pH stress
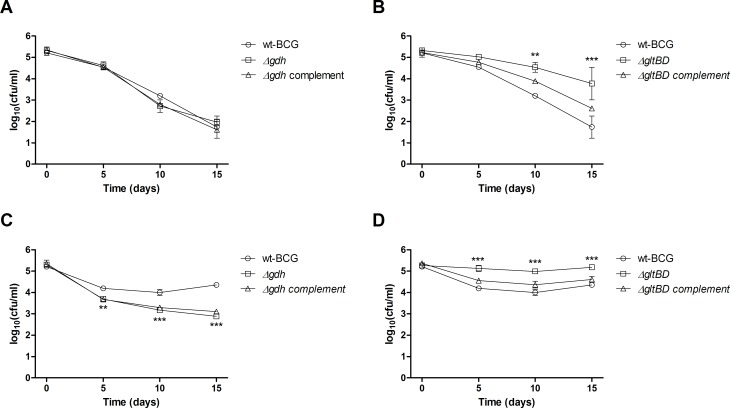
It is well documented that macrophages acidify their intracellular environment in response to invading pathogens [20,21]. It has been proposed that M. tuberculosis is able to circumvent macrophage acidification through release of ammonia, which can act as an alkalising agent in an acidic environment by sequestering hydrogen ions to produce ammonium [22,23]. Since GDH converts glutamate to ammonia, we hypothesised that GDH might also play a role in offering protection against acidic stress. While replication was observed for both the Δgdh and ΔgltBD mutants as well as the wild type strain after about 1 week of incubation in 7H9 medium of which the pH was adjusted to 4.5 with HCl, CFU enumerations for an additional week showed that replication of only the Δgdh mutant was inhibited by the acidic medium (Fig 3A). This inhibition of bacterial division was less pronounced at pH 5.5, but the replication rate of the Δgdh mutant was still reduced in comparison to the wild type strain and Δgdh complemented strain (Fig 3C). Strikingly, growth of the ΔgltBD mutant was unaffected by the low pH conditions investigated in the current study (Fig 3B and 3D). In contrast, deletion of gtlBD offered marginal protection against acidic stress at pH 4.5 (Fig 3B) and pH 5.5 (Fig 3D) and in both instances re-introduction of an intact copy of the gltBD operon restored sensitivity to wild type levels. These results indicate that degradation of glutamate through the deaminating activity of GDH offers some degree of protection against an acidic environment, probably through the release of buffering ammonia molecules into the milieu. However, our observation that a deficiency in glutamate production through GOGAT activity also confers some tolerance to acidic stress, may indicate that resistance to acidic stress may be directly correlated with intracytosolic glutamate levels, which is likely to be decreased in the ΔgltBD mutant and increased in the Δgdh mutant.
Fig 3. The effect of pH on survival of M. bovis BCG wild type, mutant (Δgdh and ΔgltBD) and complemented strains.
Exponential phase cultures were diluted to an OD600 of 0.0005 into 7H9 of which the pH was adjusted to 4.5 (A & B) or 5.5 (C & D). Symbols and error bars are means and standard errors calculated from triplicate plating obtained from two independent experiments. Data was analysed by a regular two-way ANOVA test with Bonferroni post-testing to compare mean CFU/ml of mutant (Δgdh and ΔgltBD) and wild type strains. * p < 0.05, ** p < 0.01, *** p < 0.001.
Inactivation of gdh increases sensitivity to nitrosative stress
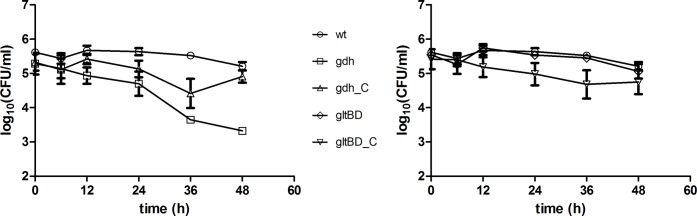
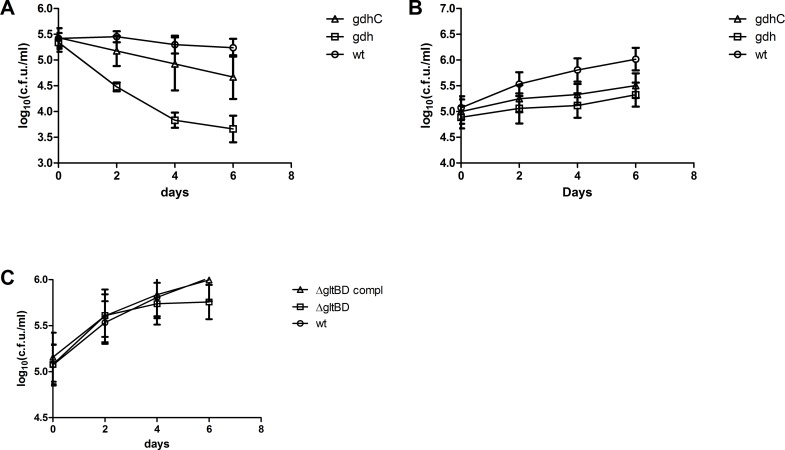
Nitric oxide (NO) is an essential component of the innate immune defence against invading pathogens and is known to play an important role in the defence of murine macrophages against M. tuberculosis infection [24]. To test the effect of nitrosative stress on M. bovis BCG and the involvement of gdh and gltBD, the wild type, mutant and complemented strains were challenged for 48 hours with 500 μM of diethelenetriamine/nitric oxide adduct (DETE/NO) in 7H9 lacking catalase. Under this in vitro nitrosative condition, division of the wild type strain was inhibited over the 48 hour period, but bacteria remained viable as no significant reduction in CFU was observed over this period (Fig 4A). However the Δgdh mutant exhibited a decline in viability, with a 2 log difference at 48 hours compared to wild type and complemented strains (Fig 4A). Interestingly, when the gdh mutant was cultivated with excess ammonium sulphate (20 mM and 30 mM) there was no decline in cell viability observed at the 48 hour time point post challenge (S1 Fig). A significant decline in viability was not observed for the ΔgltBD mutant (Fig 4B). These results indicate that the degradation of glutamate to produce 2-oxoglutarate and ammonia offers resistance to the nitrosative effect of NO.
Fig 4. The effect of diethylenetriamine/nitric oxide adduct (DETE/NO) exposure on survival of M. bovis BCG wild type, mutant (Δgdh and ΔgltBD) and complemented strains.
Exponential phase cultures of wild type, Δgdh and Δgdh complement (A) or ΔgltBD and ΔgltBD complement (B) were diluted to an OD600 of 0.0005 into 7H9 (without catalase), which was supplemented with 500 μM of diethylenetriamine/nitric oxide adduct (DETE-NO) (Sigma-Aldrich, USA) and cultured for 48 hours at 37°C without agitation. Symbols and error bars are means and standard errors calculated from three independent experiments. Data was analysed by a regular two-way ANOVA test with Bonferroni post-testing to compare mean CFU/ml of mutant (Δgdh and ΔgltBD) and wild type strains. * p < 0.05, ** p < 0.01, *** p < 0.001.
gdh is required for optimal growth in murine macrophages
In order to determine whether the contribution of gdh in M. bovis BCG resistance to acidic and nitrosative stress in vitro is actually relevant to bacterial survival inside host macrophages, we used murine macrophages as a cellular model of mycobacterial infection. As a start we studied the survival of the wild type Δgdh mutant and complemented strains in RAW 264.7 macrophages stimulated with lipopolysaccharides (LPS) from Escherichia coli, which is a macrophage cell line and represented a simple model system. While M. Bovis BCG maintained an equivalent bacterial load for the duration of infection in RAW 264.7 macrophages, Δgdh became non-viable with a nearly 2 log reduction in CFU after 6 days of infection (Fig 5A). Complementation improved viability of the mutant significantly, but not to the level seen for the wild type strain (Fig 5A). To corroborate our observation that the survival of the Δgdh mutant is reduced in macrophages, we repeated the infection experiment using the ex vivo murine bone-marrow derived macrophage (BMDM) model and including the ΔgltBD mutant and complement in the assay. In contrast to our earlier finding in RAW 264.7 macrophages, wild type M. bovis BCG growth was not suppressed in BMDM, but the replication rate of the Δgdh mutant was significantly impaired in comparison to the wild type strain (Fig 5B). Complementation of the Δgdh mutant did not restore the wild type replication rate (Fig 5B), probably due to poor expression of GDH (S1 Table). In contrast, the replication rate of the ΔgltBD mutant was not significantly different from the wild type strain in BMDM (Fig 5C). These results strongly implicate GDH in playing a substantial role in protecting infecting tubercle bacilli against host macrophage defence strategies.
Fig 5. Survival of M. bovis BCG wild type, Δgdh mutant and Δgdh complemented strains in macrophages.
RAW 264.7 macrophages (A) or BMDM (B) were infected with M. bovis BCG wild type, Δgdh mutant and Δgdh complemented strains or (C) the ΔgltBD mutant and complemented strains and the survival of the bacteria was determined by measuring CFU obtained from macrophage lysates. Means and standard errors were calculated from three independent experiments. For the BMDM infections a different mouse was used in each experiment. Data was analysed by repeated measures two-way ANOVA with Bonferroni post-testing to compare mean CFU/ml of Δgdh mutant and wild type strains. * p < 0.05, ** p < 0.01, *** p < 0.001.
Discussion
We previously reported on our success to inactivate both gdh, which encodes the primary enzyme of glutamate catabolysis, and gltBD, which encodes the primary enzyme of glutamate synthesis, in M. bovis BCG, despite their in vitro essentiality in closely related M. tuberculosis [13]. Constructing these mutants allowed us to study the roles of these genes and their protein products in nitrogen metabolism in M. bovis BCG as a model slow growing mycobacterium. We showed that gdh is required for the utilisation of glutamate as a sole nitrogen source and when the amino acids asparagine and aspartate are present in excess concentrations [13]. On the other hand we found that M. bovis BCG became glutamate auxotrophic when gltBD was inactivated [13]. This glutamate auxotrophy could however be rescued by supplementation with amino acids such as aspartate, glutamine and asparagine [13].
In addition to a role as a precursor or nitrogen donor in the biosynthesis of a range of nitrogenous molecules [25,26], glutamate may be involved to a great extent in other functions that may be critical to the intracellular lifestyle and pathogenesis caused by slow growing mycobacterial pathogens such as M. tuberculosis. These include a source of carbon in its carbon backbone structure which may be freed as a 2-oxoglutarate molecule (a TCA cycle intermediate), a function in acid resistance [27,28], osmoprotection [29] and as a compatible solute [30]. Therefore, we extended our study of glutamate metabolism here by using the same mutants and in vitro conditions that mimic some of the bactericidal mechanisms employed by macrophage cells as well as two macrophage infection models to further delineate the role of glutamate in the pathogenicity of slow growing mycobacteria. We show that gdh is indispensable for the utilisation of glutamate as a major carbon source even in the presence of cholesterol, a neutral lipid which was previously shown to constitute a major source of carbon for M. tuberculosis in a chronic infection mouse model [19,31]. Mycobacterial central metabolism is different when cultured in medium containing in addition to asparagine, cholesterol or glycerol as major carbon sources, with central metabolites such as succinate, fumarate and malate accumulating only in the medium containing cholesterol [32]. Interestingly, these methylcitrate cycle intermediates are also downstream products of glutamate catabolism through the release of 2-oxoglutarate. It could be speculated that the poor growth of the Δgdh mutant in medium containing both cholesterol and glutamate as carbon sources is a result of deregulation associated with 2-oxoglutarate metabolism, which is not ameliorated by supplementation with cholesterol, however a deeper investigation is warranted to understand the interplay between glutamate and cholesterol catabolism. We also show that gdh is required for survival in a low pH environment as well as under nitrosative stress, two defence mechanisms employed by macrophages to destroy phagocytosed microbes [20,21]. Under none of the tested conditions could we show a critical involvement for gltBD. Finally, we show that gdh is required for optimal survival of M. bovis BCG in murine macrophages.
Compellingly, our results are in agreement with recent studies into the uptake and metabolism of the amino acids glutamine, asparagine and aspartate, which are easily converted to glutamate, and their involvement in survival and pathogenicity of M. tuberculosis. Especially the highly expressed essential ammonium assimilating enzyme glutamine synthetase (GS), has garnered a lot of attention for its involvement in M. tuberculosis virulence [22,33–37]. However, this enzyme is mostly important under conditions when amino acids are not available as sources of nitrogen and only ammonium is available for the de novo synthesis of glutamine [22,33,38]. It was more recently found that a putative asparagine/aspartate transporter (AnsP1) is able to transport aspartate across the cell envelope and deletion of ansP1 resulted in impaired virulence of M. tuberculosis in a mouse model, clearly demonstrating that the assimilation of aspartate by M. tuberculosis is required for virulence [39]. The same group inactivated ansP2, which encodes another permease transporting asparagine across the cell envelope, but found this gene to be non-essential to virulence in mice [40]. However, when they inactivated ansA, which encodes an asparaginase, which readily converts asparagine to aspartate and ammonia a growth defect was observed in mice [40]. Interestingly, AnsA is secreted and it was speculated by the authors that the conversion of asparagine to aspartate is necessary for virulence due to the release of ammonia to the M. tuberculosis extracellular environment [40]. Ammonia may alkalise the phagosome and circumvent acidification and maturation, two crucial processes of macrophage defence against intracellular infection [23]. Through this mechanism, M. tuberculosis can initiate an efficient response against the host defences while simultaneously gaining nutrition in the form of nitrogen and carbon through the assimilation of aspartate. In M. tuberculosis, aspartate could be transaminated by at least three putative aspartate aminotransferases (AspB, AspC and Rv3722c) to produce glutamate, which in turn could be deaminated by GDH to produce free ammonia and 2-oxoglutarate.
Our observation that gdh is also an important factor in protection against nitrosative stress was unanticipated. It has been found that high levels of intracellular ammonia may induce the expression of enzymes involved in protection against oxidative stress [40,41–43]. In addition, it was more recently found that 2-hydroxy-3-oxoadipate synthase (HOAS) of the KDH complex in M. tuberculosis forms part of a four component peroxidase system which uses 2-oxoglutarate as an electron donor in a reductase reaction [44], which might also implicate 2-oxoglutarate in protection against nitrosative stress as a product of glutamate degradation through GDH activity.
Hence, we propose the following model, depicted in Fig 6, whereby GDH plays a critical role in survival of pathogenic mycobacteria inside macrophages: (i) as an effector of nitrogen and (ii) carbon assimilation by degrading glutamate to release ammonium and 2-oxoglutarate, respectively, (iii) by protecting against acidic stress through release of buffering ammonia from glutamate degradation, and (iv) by protecting against nitrosative stress through a yet unknown mechanism which may involve priming of oxidative stress mechanisms by sustaining cytosolic ammonia levels or through the release of the 2-oxoglutarate backbone of the glutamate molecule.
Fig 6. The role(s) of enzymes related to central mycobacterial glutamate metabolism and metabolites in the resistance against acid and nitrosative stress.
Glutamates is an important nitrogen source (i), carbon source (ii) protectant against acidic stress (iii) as well as against nitrosative stress (iv). Metabolites shown in red are responsible for resistance against either nitrosative stress or acidic stress. Enzymes marked with circles have been demonstrated to be required for resistance against cellular stress.
Conclusion
Unexpectedly, our data support a much more important role for GDH than GOGAT in the intracellular lifestyle of slow growing mycobacteria, such as M. tuberculosis. The unique properties of L180 GDH compared to other characterised GDHs, including a very large subunit size, exclusive NAD+ co-enzyme specificity, and exclusive distribution among bacteria may have positive implications for the potential of L180 GDH as a specific anti-TB drug target [45]. A protein BLAST of M. tuberculosis gdh against the genomes for common intestinal bacterial genera, including Bacteroides, Enterococcus, Escherichia, Klebsiella, Staphylococcus, Lactobacillus and Clostridium delivered no homologues, which may further qualify L180 GDH as a specific anti-TB drug target (query coverage < 40 for only two Lactobacilli and one Bacteroides, http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Supporting Information
All strains were cultivated in excess ammonium sulphate (20 mM or 30 mM). Cells were washed 6 times with PBS prior to DETE/NO challenge and diluted to an OD600 0.0005. Percentages were calculated from CFU enumerations. Data was obtained from three independent experiments.
(TIF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
The authors would like to thank Carine Sao Emani, Monique Williams, and Carrie Kirsten for insightful discussions about the work presented in this report as well as Jomien Mouton, Ray-Dean Pietersen and Bienyameen Baker for technical advices with macrophages experiments.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
JG would like to acknowledge the NRF/MRC for financial assistance through the Allied Health Scholarship. AV would like to acknowledge the Harry Crossley Foundation, the NRF-DAAD and L’Infectionpôle Sud for financial assistance. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO | Global tuberculosis report 2014. In: WHO Available: http://www.who.int/tb/publications/global_report/en/.
- 2.Gouzy A, Poquet Y, Neyrolles O. Nitrogen metabolism in Mycobacterium tuberculosis physiology and virulence. Nat Rev Microbiol. 2014;12: 729–737. 10.1038/nrmicro3349 [DOI] [PubMed] [Google Scholar]
- 3.Rhee KY, de Carvalho LPS, Bryk R, Ehrt S, Marrero J, Park SW, et al. Central carbon metabolism in Mycobacterium tuberculosis: an unexpected frontier. Trends Microbiol. 2011;19: 307–314. 10.1016/j.tim.2011.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cowley S, Ko M, Pick N, Chow R, Downing KJ, Gordhan BG, et al. The Mycobacterium tuberculosis protein serine/threonine kinase PknG is linked to cellular glutamate/glutamine levels and is important for growth in vivo. Mol Microbiol. 2004;52: 1691–702. [DOI] [PubMed] [Google Scholar]
- 5.Schultz C, Niebisch A, Gebel L, Bott M. Glutamate production by Corynebacterium glutamicum: dependence on the oxoglutarate dehydrogenase inhibitor protein OdhI and protein kinase PknG. Appl Microbiol Biotechnol. 2007;76: 691–700. [DOI] [PubMed] [Google Scholar]
- 6.O’Hare HM, Durán R, Cerveñansky C, Bellinzoni M, Wehenkel AM, Pritsch O, et al. Regulation of glutamate metabolism by protein kinases in mycobacteria. Mol Microbiol. 2008;70: 1408–23. 10.1111/j.1365-2958.2008.06489.x [DOI] [PubMed] [Google Scholar]
- 7.Nott TJ, Kelly G, Stach L, Li J, Westcott S, Patel D, et al. An intramolecular switch regulates phosphoindependent FHA domain interactions in Mycobacterium tuberculosis. Sci Signal. 2009;2: ra12 10.1126/scisignal.2000212 [DOI] [PubMed] [Google Scholar]
- 8.Ventura M, Rieck B, Boldrin F, Degiacomi G, Bellinzoni M, Barilone N, et al. GarA is an essential regulator of metabolism in Mycobacterium tuberculosis. Mol Microbiol. 2013; n/a–n/a. 10.1111/mmi.12368 [DOI] [PubMed] [Google Scholar]
- 9.Owen OE, Kalhan SC, Hanson RW. The Key Role of Anaplerosis and Cataplerosis for Citric Acid Cycle Function. J Biol Chem. 2002;277: 30409–30412. 10.1074/jbc.R200006200 [DOI] [PubMed] [Google Scholar]
- 10.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48: 77–84. [DOI] [PubMed] [Google Scholar]
- 11.Griffin JE, Gawronski JD, Dejesus MA, Ioerger TR, Akerley BJ, Sassetti CM. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 2011;7: e1002251 10.1371/journal.ppat.1002251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeJesus MA, Zhang YJ, Sassetti CM, Rubin EJ, Sacchettini JC, Ioerger TR. Bayesian analysis of gene essentiality based on sequencing of transposon insertion libraries. Bioinformatics. 2013;29: 695–703. 10.1093/bioinformatics/btt043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Viljoen AJ, Kirsten CJ, Baker B, van Helden PD, Wiid IJF. The Role of Glutamine Oxoglutarate Aminotransferase and Glutamate Dehydrogenase in Nitrogen Metabolism in Mycobacterium bovis BCG. PLoS ONE. 2013;8 10.1371/journal.pone.0084452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung YW, Tanner JA. Targeting glutamate synthase for tuberculosis drug development. Hong Kong Med J Xianggang Yi Xue Za Zhi Hong Kong Acad Med. 2011;17 Suppl 2: 32–34. [PubMed] [Google Scholar]
- 15.Rengarajan J, Bloom BR, Rubin EJ. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc Natl Acad Sci U S A. 2005;102: 8327–8332. 10.1073/pnas.0503272102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muñoz-Elías EJ, McKinney JD. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat Med. 2005;11: 638–644. 10.1038/nm1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edson NL. The intermediary metabolism of the mycobacteria. Bacteriol Rev. 1951;15: 147–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyon RH, Rogers P, Hall WH, Lichstein HC. Inducible Glutamate Transport in Mycobacteria and Its Relation to Glutamate Oxidation1. J Bacteriol. 1967;94: 92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pandey AK, Sassetti CM. Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci U S A. 2008;105: 4376–4380. 10.1073/pnas.0711159105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lundborg M, Falk R, Johansson A, Kreyling W, Camner P. Phagolysosomal pH and dissolution of cobalt oxide particles by alveolar macrophages. Environ Health Perspect. 1992;97: 153–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nyberg K, Johansson U, Johansson A, Camner P. Phagolysosomal pH in alveolar macrophages. Environ Health Perspect. 1992;97: 149–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tullius MV, Harth G, Horwitz MA. Glutamine synthetase GlnA1 is essential for growth of Mycobacterium tuberculosis in human THP-1 macrophages and guinea pigs. Infect Immun. 2003;71: 3927–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gordon AH, Hart PD, Young MR. Ammonia inhibits phagosome-lysosome fusion in macrophages. Nature. 1980;286: 79–80. [DOI] [PubMed] [Google Scholar]
- 24.Yang C-S, Yuk J-M, Jo E-K. The Role of Nitric Oxide in Mycobacterial Infections. Immune Netw Off J Korean Soc Immunol Biol Response Modif. 2009;9: 46–52. 10.4110/in.2009.9.2.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40: D109–114. 10.1093/nar/gkr988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richard H, Foster JW. Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J Bacteriol. 2004;186: 6032–6041. 10.1128/JB.186.18.6032-6041.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bearson BL, Lee IS, Casey TA. Escherichia coli O157 : H7 glutamate- and arginine-dependent acid-resistance systems protect against oxidative stress during extreme acid challenge. Microbiol Read Engl. 2009;155: 805–812. 10.1099/mic.0.022905-0 [DOI] [PubMed] [Google Scholar]
- 29.Goude R, Renaud S, Bonnassie S, Bernard T, Blanco C. Glutamine, Glutamate, and α-Glucosylglycerate Are the Major Osmotic Solutes Accumulated by Erwinia chrysanthemi Strain 3937. Appl Environ Microbiol. 2004;70: 6535–6541. 10.1128/AEM.70.11.6535-6541.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saum SH, Sydow JF, Palm P, Pfeiffer F, Oesterhelt D, Müller V. Biochemical and molecular characterization of the biosynthesis of glutamine and glutamate, two major compatible solutes in the moderately halophilic bacterium Halobacillus halophilus. J Bacteriol. 2006;188: 6808–6815. 10.1128/JB.00781-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brzostek A, Pawelczyk J, Rumijowska-Galewicz A, Dziadek B, Dziadek J. Mycobacterium tuberculosis Is Able To Accumulate and Utilize Cholesterol. J Bacteriol. 2009;191: 6584–6591. 10.1128/JB.00488-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Griffin JE, Pandey AK, Gilmore SA, Mizrahi V, Mckinney JD, Bertozzi CR, et al. Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chem Biol. 2012;19: 218–227. 10.1016/j.chembiol.2011.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harth G, Horwitz MA. Inhibition of Mycobacterium tuberculosis glutamine synthetase as a novel antibiotic strategy against tuberculosis: demonstration of efficacy in vivo. Infect Immun. 2003;71: 456–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nilsson MT, Krajewski WW, Yellagunda S, Prabhumurthy S, Chamarahally GN, Siddamadappa C, et al. Structural Basis for the Inhibition of Mycobacterium tuberculosis Glutamine Synthetase by Novel ATP-Competitive Inhibitors. J Mol Biol. 2009;393: 504–513. 10.1016/j.jmb.2009.08.028 [DOI] [PubMed] [Google Scholar]
- 35.Gising J, Nilsson MT, Odell LR, Yahiaoui S, Lindh M, Iyer H, et al. Trisubstituted Imidazoles as Mycobacterium tuberculosis Glutamine Synthetase Inhibitors. J Med Chem. 2012;55: 2894–2898. 10.1021/jm201212h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nordqvist A, Nilsson MT, Lagerlund O, Muthas D, Gising J, Yahiaoui S, et al. Synthesis, biological evaluation and X-ray crystallographic studies of imidazo[1,2-a]pyridine-based Mycobacterium tuberculosis glutamine synthetase inhibitors. MedChemComm. 2012;3: 620–626. 10.1039/C2MD00310D [DOI] [Google Scholar]
- 37.Odell LR, Nilsson MT, Gising J, Lagerlund O, Muthas D, Nordqvist A, et al. Functionalized 3-amino-imidazo[1,2-a]pyridines: A novel class of drug-like Mycobacterium tuberculosis glutamine synthetase inhibitors. Bioorg Med Chem Lett. 2009;19: 4790–4793. 10.1016/j.bmcl.2009.06.045 [DOI] [PubMed] [Google Scholar]
- 38.Harth G, Horwitz MA. An inhibitor of exported Mycobacterium tuberculosis glutamine synthetase selectively blocks the growth of pathogenic mycobacteria in axenic culture and in human monocytes: extracellular proteins as potential novel drug targets. J Exp Med. 1999;189: 1425–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gouzy A, Larrouy-Maumus G, Wu T-D, Peixoto A, Levillain F, Lugo-Villarino G, et al. Mycobacterium tuberculosis nitrogen assimilation and host colonization require aspartate. Nat Chem Biol. 2013;9: 674–676. 10.1038/nchembio.1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gouzy A, Larrouy-Maumus G, Bottai D, Levillain F, Dumas A, Wallach JB, et al. Mycobacterium tuberculosis Exploits Asparagine to Assimilate Nitrogen and Resist Acid Stress during Infection. PLoS Pathog. 2014;10: e1003928 10.1371/journal.ppat.1003928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang W, Xiang F, Liang L, Yang Z. Toxicity of Ammonia and Its Effects on Oxidative Stress Mechanisms of Juvenile Crucian Carp (Carassius auratus). J Freshw Ecol. 2010;25: 297–302. 10.1080/02705060.2010.9665080 [DOI] [Google Scholar]
- 42.Kosenko E, Kaminsky M, Kaminsky A, Valencia M, Lee L, Hermenegildo C, et al. Superoxide Production and Antioxidant Enzymes in Ammonia Intoxication in Rats. Free Radic Res. 1997;27: 637–644. 10.3109/10715769709097867 [DOI] [PubMed] [Google Scholar]
- 43.Sinha AK, Zinta G, AbdElgawad H, Asard H, Blust R, De Boeck G. High environmental ammonia elicits differential oxidative stress and antioxidant responses in five different organs of a model estuarine teleost (Dicentrarchus labrax). Comp Biochem Physiol Toxicol Pharmacol CBP. 2015;174–175: 21–31. 10.1016/j.cbpc.2015.06.002 [DOI] [PubMed] [Google Scholar]
- 44.Maksymiuk C, Balakrishnan A, Bryk R, Rhee KY, Nathan CF. E1 of α-ketoglutarate dehydrogenase defends Mycobacterium tuberculosis against glutamate anaplerosis and nitroxidative stress. Proc Natl Acad Sci. 2015; 201510932 10.1073/pnas.1510932112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miñambres B, Olivera ER, Jensen RA, Luengo JM. A new class of glutamate dehydrogenases (GDH). Biochemical and genetic characterization of the first member, the AMP-requiring NAD-specific GDH of Streptomyces clavuligerus. J Biol Chem. 2000;275: 39529–42. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All strains were cultivated in excess ammonium sulphate (20 mM or 30 mM). Cells were washed 6 times with PBS prior to DETE/NO challenge and diluted to an OD600 0.0005. Percentages were calculated from CFU enumerations. Data was obtained from three independent experiments.
(TIF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.