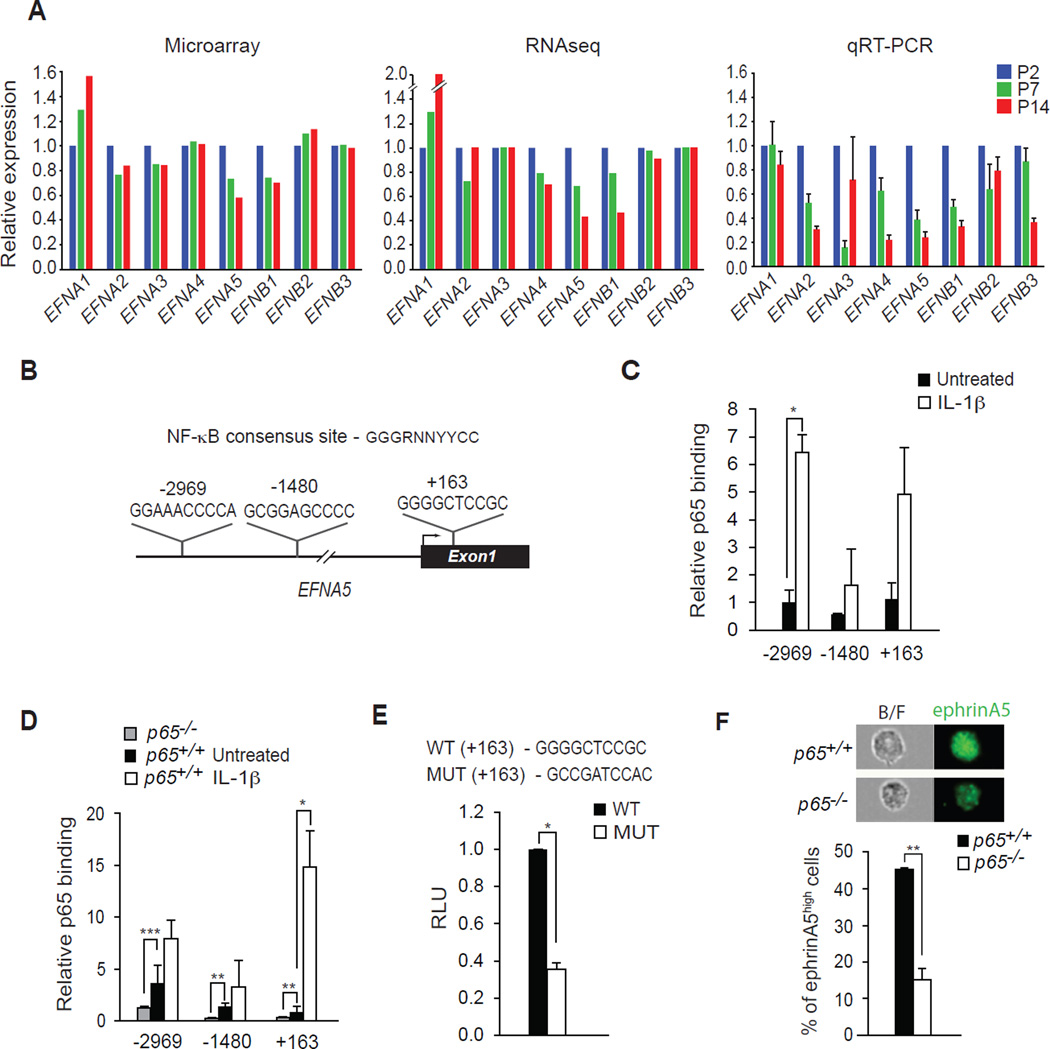
Figure 3. EFNA5 is Transcriptionally Regulated by NF-κB.
(A) Microarray, RNAseq, and quantitative RT-PCR were performed probing for the expression of ephrin genes from skeletal muscles at P2, P7, and P14. (B) A schematic map of the 5’ region of the EFNA5 gene and locations of conserved NF-κB DNA binding sites at −2969, −1480, and +163 relative to the transcriptional start site. (C) ChIP assays for p65 binding to the EFNA5 promoter in NG2+ cells untreated and treated with 10 ng/ml of IL-1β. (D) ChIP assays for p65 binding to the EFNA5 promoter in p65+/+ MEFs untreated and treated with 10 ng/ml of IL-1β, compared with untreated p65−/− MEFs *** p < 0.001. (E) A portion of EFNA5 gene containing the +163 binding site of NF-κB was cloned into a luciferase reporter construct. Luciferase activity was measured following the transfection of an EFNA5 promoter reporter construct in MEFs, and compared to activity generated from an EFNA5 promoter reporter construct containing a mutation in the +163 NF-κB binding site. (F) Representative flow images of ephrinA5 from p65+/+ and p65−/− NG2 cells (top). Percentage of ephrinA5high NG2 cells isolated from p65+/+ and p65−/− cells (bottom). RLU: Relative Luciferase Units. B/F: bright field image. *p < 0.05, **p < 0.01 by the student’s t-test. See accompanying Figure S3. Error bars, SEM