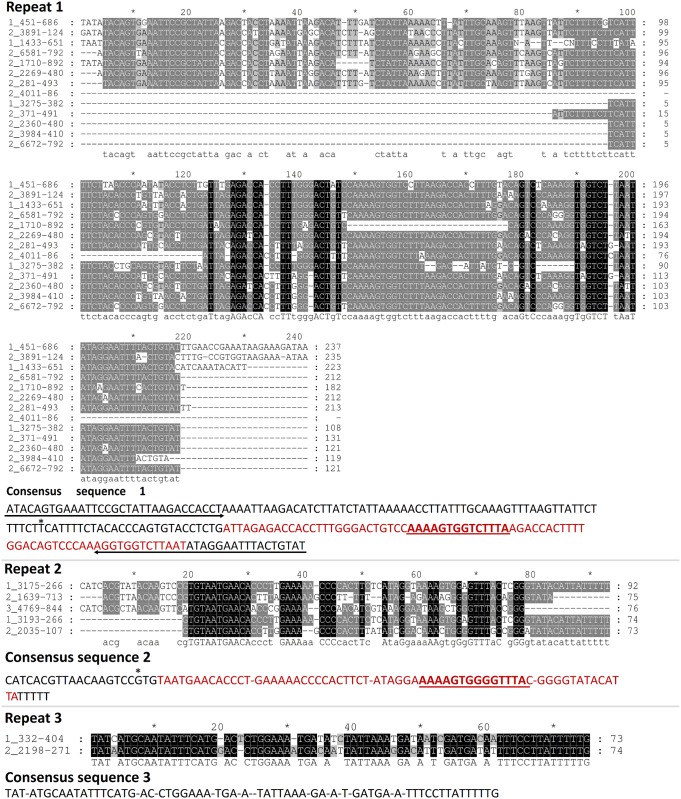
Fig 5. Alignment of repetitive segments found in L. pectinata’s Hbs genes.
The first number of each sequence is 1, 2 and 3 for HbI, HbII and HbIII, respectively. The following number indicates the region of each sequence in its respective gene. Three alignments, named repeat 1, 2 and 3 were generated using Clustal W [23] and visualized and formatted with GeneDoc [35]. For each repeat, a consensus sequence was derived according to the majority principle and is shown below each alignment. In consensus sequence 1, arrows specified imperfect inverted repeats found at the flanking regions of this sequence, bases in red indicate the self-complementary segment and the asterisk indicates the start of the truncated sequences. Consensus sequence 2 has a self-complementary segment represented with letters in red. It also has a short segment in common with consensus sequence 1 which is indicated as underlined letters. Consensus sequence 3 has few similarities with other consensus sequences.