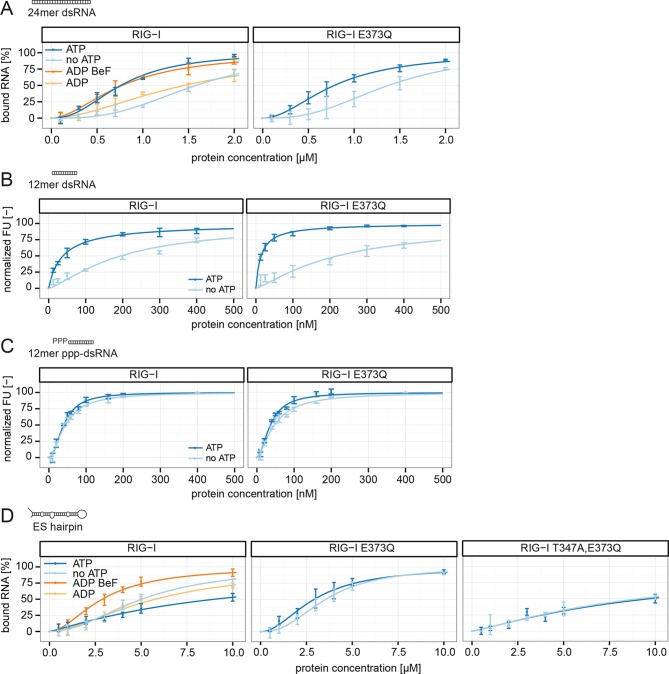
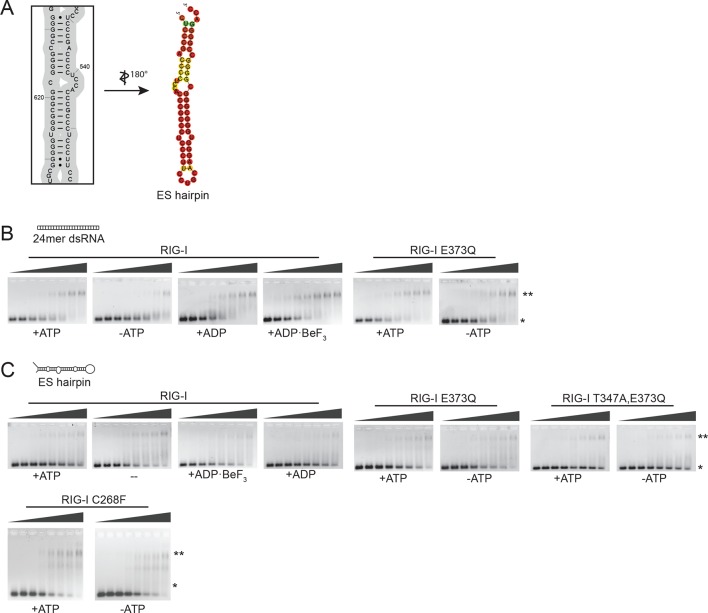
Figure 4. RIG-I’s ATP hydrolysis enhances RNA end recognition and removes RIG-I from RNA stems.
(A) Quantification of electrophoretic mobility shift assays of RIG-I or RIG-I E373Q incubated with 24mer dsRNA in presence or absence of ATP, ADP or ADP·BeF3 (compare with Figure 4—Figure supplement 1B). (B) Fluorescence anisotropy changes measured by titrating RIG-I or RIG-I E373Q in presence or absence of ATP into solutions containing fluorescently labeled 12mer dsRNA. (C) Fluorescence anisotropy changes measured by titrating RIG-I or RIG-I E373Q in presence or absence of ATP into solutions containing fluorescently labeled 12mer ppp-dsRNA. (D) Quantification of electrophoretic mobility shift assays of RIG-I, RIG-I E373Q or RIG-I T347A, E373Q incubated with an RNA hairpin derived from helix A of the human ribosome expansion segment 7L (ES hairpin) in presence or absence of ATP, ADP or ADP·BeF3 (compare with Figure 4—Figure supplement 1C). All binding curves were fitted using the LL.2 function of the R drc package (Cedergreen et al., 2005). n=3-6, error bars represent mean values ± standard deviation.