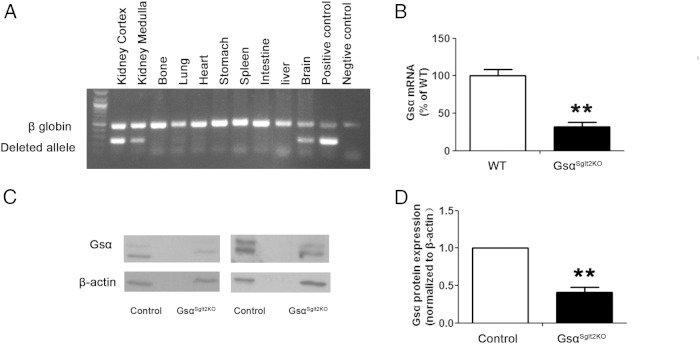
Figure 2.
GsαSglt2KO show significantly reduced Gsα expression in PT. A, A duplex PCR was used to amplify the deleted Gnas exon 1 allele (deleted allele) together with a portion of the gene encoding β-globin (β-globin) in genomic DNA (35 cycles of amplification was used). The positive control was from the tail of a mouse in which the paternal Gnas exon 1 was deleted universally through the same Cre/lox approach as in GsαSglt2KO. The other lanes were from different tissues of GsαSglt2KO mice (representative of 2 independent experiments). B, Gsα mRNA was analyzed by qRT-PCR using RNA isolated from proximal tubules by LCM. For LCM, the tubules were marked in the double mutant offspring of Sglt2-Cre and RosatdTomato mice, both of which were also heterozygous for the floxed exon 1 allele. Double transgenic offspring carrying homozygous floxed alleles (KO) was compared with double transgenic offspring carrying no floxed alleles (WT). n = 3 per group. **, P < .01. C, Gsα protein level was examined in proximal tubule enriched renal cortices by Western blotting (representative data from 3 independent experiments). D, Densitometry analysis showing the reduction in Gsα protein level in GsαSglt2KO mice compared with control littermates (results from 3 independent experiments). n = 4 per genotype; **, P < .01.