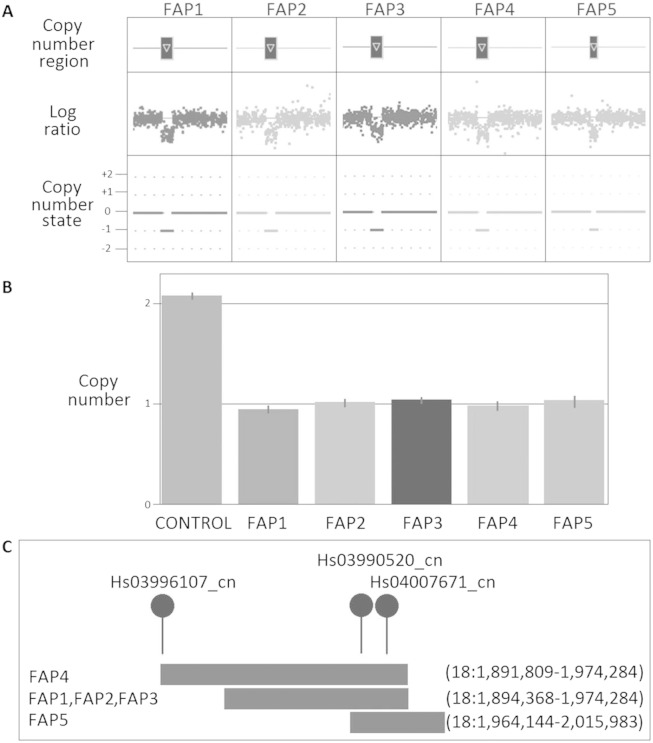
Fig. 3.
CNV results for 18p11.32 deletion in the FAP patients (FAP1, FAP2, FAP3, FAP4 and FAP5). (A) CNV profile from Cyto2.7M array data using ChAS noting the defined CN region (dark box above gene representing the CN deletion) in relation to the log ratio plot (relative fluorescence of each probe, dot, on the array showing a decrease in fluorescence indicating a loss in genomic material) and the CN state (0 = normal two copies present, + 1 = one extra copy, + 2 = two extra copies, -1 = one less copy and -2 = two less copies); (B) Validation using TaqMan CN assay showing results for assay Hs03990520_cn noting the normal two copies of this region identified in the control (CON2), confirmation of the aberrant one copy in all affected FAP patients and the error bars associated with the three technical repeats for each sample; and (C) Location of CN deletions with respect to each other and the TaqMan CN assays used in validating the variants.