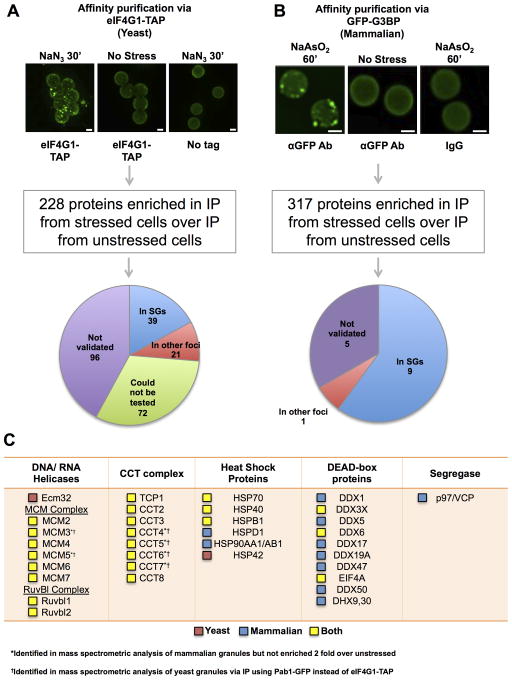
Figure 3. Mass Spectrometric Analysis of Stress Granule Cores.
(A) Dynabeads bound to immunopurified yeast stress granule cores are shown. Pie chart shows GFP-tagged proteins localized to stress granules by overlap with Ded1-mCherry (see Supplemental Experimental Procedures). (B) Stress granule core enriched fraction from GFP-G3BP U-2 OS cells incubated with αGFP antibody, immunopurified using Protein A Dynabeads. Picture shows isolated granule cores bound to Dynabeads. Pie chart shows result of testing the co-localization of 15 putative stress granule proteins with GFP-G3BP by IF. (A) and (B) Scale bars are 2μm. (C) Table showing various conserved ATPases in stress granules. Red box indicates protein present only in the yeast granule proteome, blue indicates protein present only in the mammalian granule proteome and yellow indicates protein is conserved in both proteomes. See also Figure S4 and Tables S1, S2, S3.