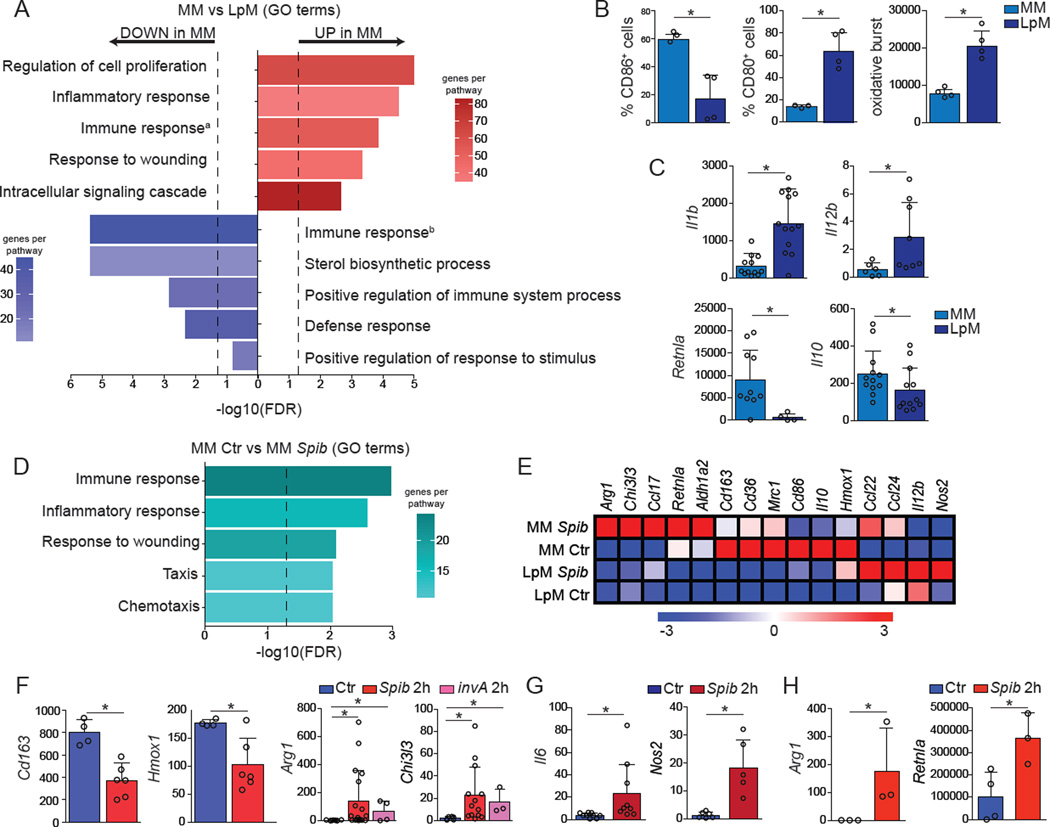
Figure 2. MMs and LpMs Exhibit Distinct Gene Expression Signatures.
(A–G) MMs and LpMs were isolated from the small intestine muscularis externa and mucosal layers, respectively, of WT mice and sorted as live (Aqua−) CD45+Lin−MHCII+F4/80+CD11B+ CD11C+CD103−. (A) Annotated gene ontology (GO) biological processes were assigned to genes differentially expressed by MMs when compared to LpMs, determined by RNA-seq. (B) Flow cytometry analysis of CD80 and CD86 expression and oxidative burst by MMs and LpMs. (C) Expression of mRNA for Il1b, Il12b, Retnla, and Il10 by MMs and LpMs determined by qPCR, presented relative to housekeeping gene Rpl32 expression. (D) Annotated GO biological processes were assigned to genes differentially expressed by MMs 2 h post intragastric exposure to Spib when compared to MMs isolated from naïve mice, determined by RNA-seq. (E) Heat map of representative M1– and M2–related gene were assigned to genes differentially expressed by MMs and LpMs 2 h post intragastric exposure to Spib, determined by RNA-seq. (F,G) Expression of mRNA for various genes by (F) MMs and (G) LpMs 2 h post intragastric exposure to Spib or invA Salmonella Typhimurium mutant strains determined by qPCR, presented relative to Rpl32 expression. (H) Expression of polysome-associated mRNA from small intestine muscularis of Lyz2 mice 2 h post intragastric exposure to Spib determined by qPCR, presented relative to Gapdh. (A, D and E), n = 2 per condition; (B), data are representative of at least 2 independent experiments, n = 4; (C), n = 6–12, pooled data; (F and G), n = 4–13; pooled data; (H), n= 3 per condition. Data were analyzed by unpaired T-test and are shown as average±SD, *p ≤ 0.05. See also Table S1, worksheets a–c). See also Figure S2.