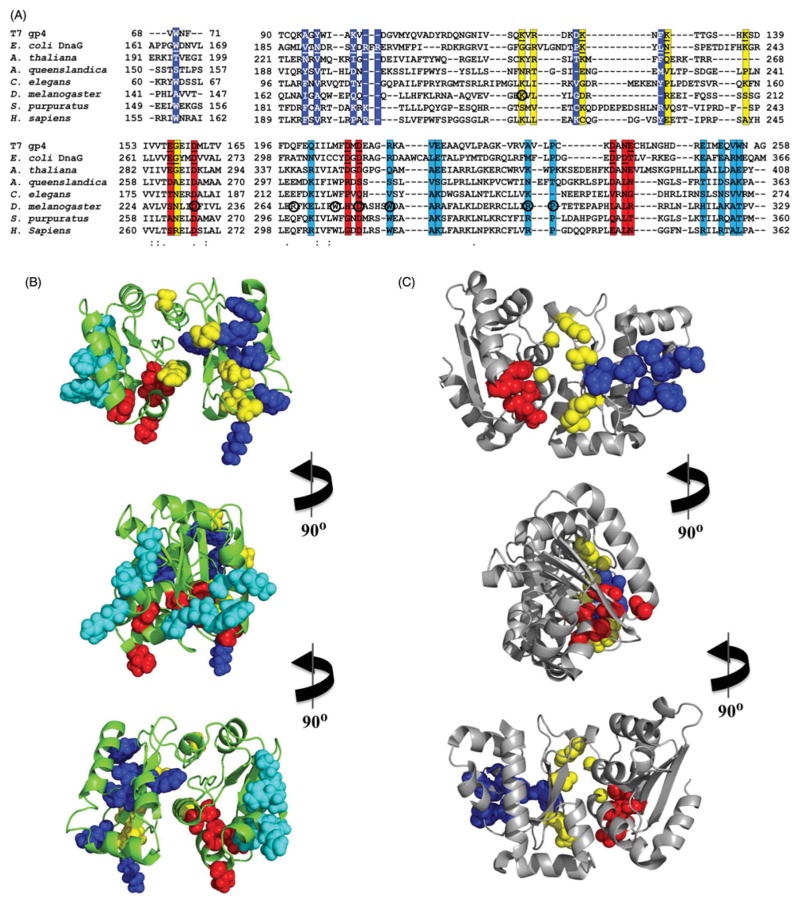
Figure 4.
Mapping of conserved amino acid residues and human pathogenic mutations in the human RPD model. (A) Amino acid sequence alignment among T7 gp4, E. coli DnaG and selected mtDNA helicases, showing important residues for ssDNA (highlighted in blue), NTP-(yellow) and Mg2+-binding (red), and human disease mutations (cyan). The residues in T7 gp4 and bacterial DnaG that were shown experimentally to bind ssDNA, NTP or Mg2+ (Corn et al., 2008; Kato et al., 2003; Keck et al., 2000; Lee & Richardson, 2001; Rymer et al., 2012) are underlined, and the residues in the D. melanogaster mtDNA helicase that have been mutated in S2 cells (Matsushima & Kaguni, 2009) are circled. These residues have been positioned on the human RPD model (B) and in the E. coli RPD structure (PDB: 1DDE, (Keck et al., 2000)) (C) to illustrate (1) that the homologous residues in the human RPD, important for primase activity in DnaG, are displaced from an active configuration and (2) that the human disease mutations (Appendix Table 1) are clustered in a region of the TOPRIM fold of the human RPD (see color version of this figure at www.informahealthcare.com/bmg).