Figure 7. Single-cell regulation of Nrxn1 ss4 selection is dependent on brain region.
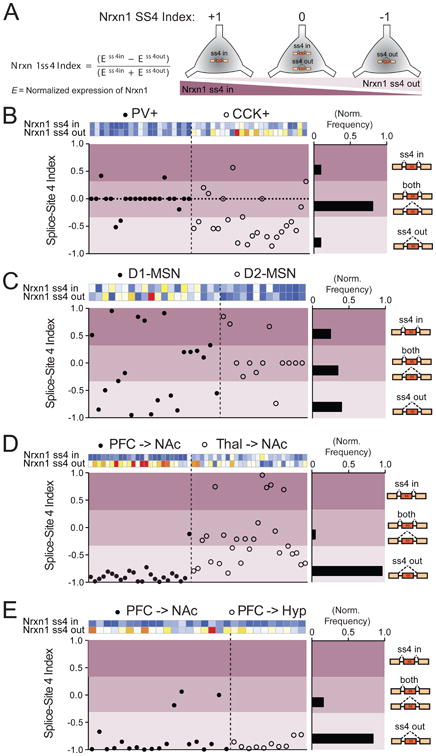
(A) Description of Nrxn1 splice-site 4 index, which assesses exclusive presence of single splice isoforms or coexpression of Nrxn1ss4-IN and Nrxn1ss4-OUT transcripts. An index=0 represents roughly equal normalized expression values for both Nrx1ss4-IN and Nrxn1ss4-OUT probes.
(B-E) Plot of splice-site 4 index for all single neurons collected in hippocampal interneuron (B, n=45 cells), NAc MSN (C, n=33 cells), NAc-projecting (D, n=51 cells), and divergent PFC projection (E, n=29 cells) experiments. Each plot shows the heat maps of both Nrx1ss4 probes for each cell, with the ss4 index plotted below. Splice-site territories were arbitrarily subdivided into ss4 in (index=0.33-1.00), both (index=-0.33– 0.33) and ss4 out (index=-1.0– -0.33) and normalized frequencies were calculated for each region (right histograms).
