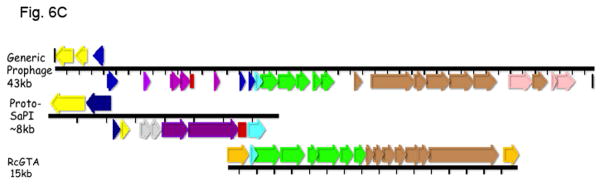
Fig. 6. A. Genomes of lactococcal PICIs.
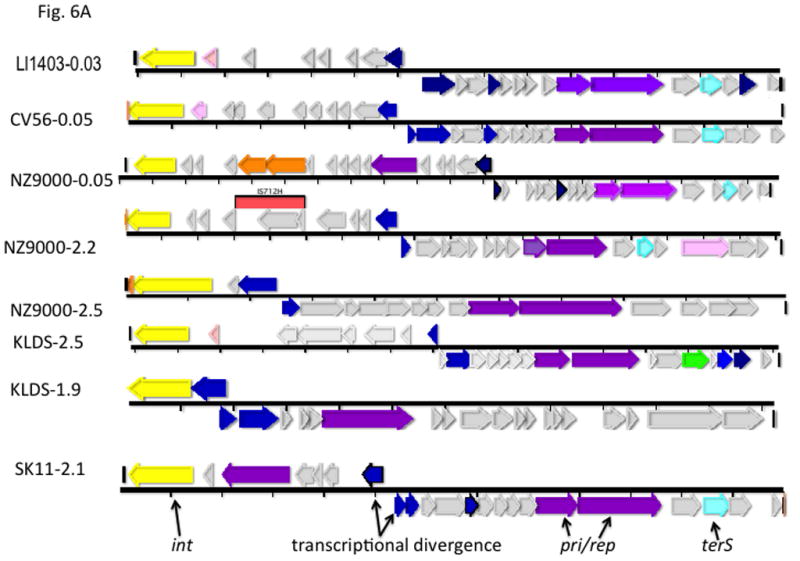
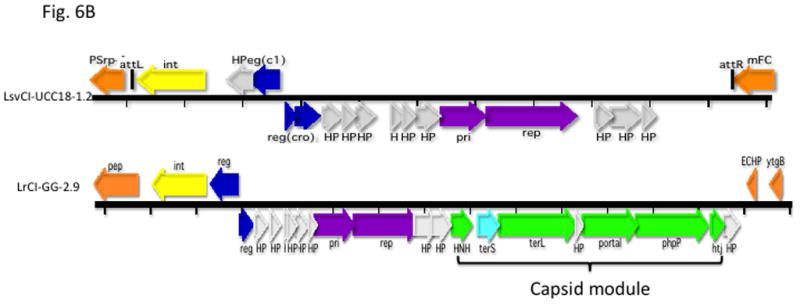
Gene coloring is the same as in Fig. 1 with the addition of green for procapsid and light red-orange for the transposon in the NZ9000(0.03) element. Numbers after the strain names represent approximate genome locations. B. Genomes of phage-related elements from lactobacilli. Color coding is as in Fig. 1, with the addition of green for capsid morphogenesis. Annotations are from GenBank. Abbreviations in addition to those in Fig. 1: LsvCI-UC1118 – L. salivarius UCC118; PSrp – ribosome-associated protein; reg (c1) –λc1-like repressor; reg (cro) - λcro-like regulator; mFC – comF protein 3; LrCI-GG – L. rhamnosus GG; pep – phosphomonomutase-like protein; reg – regulation; HNH – HNH nuclease; terL – terminase large subunit; portal – portal protein; php – prohead protease; htj – head-tail joining protein. C. Hypothetical origins of proto-SaPI and GTA. At top is shown the genome of a generic prophage that could be from either a Gram+ or a Gram- organism. Below is shown a possible “proto-SaPI” consisting of the 5′ region of the prophage from int to terS. Below the “proto-SaPI” is shown the RcGTA, consisting of most of the 3′ region of the prophage from terS to the tape measure gene, but lacking the extreme 3′ end. Gene annotations are from the KEGG orthology lists [43] or from GenBank. Colors: yellow – int/xis; blue – regulatory; purple – replication; aqua – terS; green – capsid morphogenesis; - brown – tail morphogenesis; pink – lysis; orange –flanking genes; red box – replication origin.