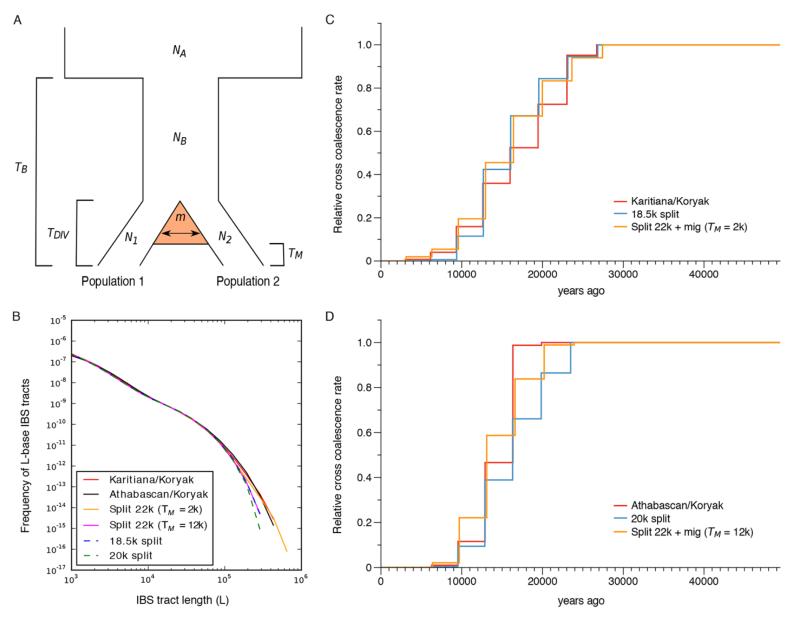
Fig. 2. Divergence estimates between Native Americans and Siberian Koryak.
(A) The demographic model used allows for continuous gene flow between populations 1 and 2, starting from the time TDIV of divergence and ending at TM. The backward probability of migration per individual per generation is denoted by m. The bottleneck at TB captures the out-of-Africa event. (B) The red and black solid curves depict empirical distributions of IBS tracts shared between Karitiana-Koryak and Athabascan-Koryak, respectively. The orange, pink, dashed blue and dashed green curves depict IBS tracts shared between the two population pairs, simulated under two demographic models based on results from diCal2.0. Overall, for Karitiana-Koryak and Athabascan-Koryak, the migration scenarios (orange and pink, respectively) match the empirical curves (red and black, respectively) better than the clean split scenarios (dashed blue and dashed green, respectively), with more long IBS tracts showing evidence of recent common ancestry between Koryaks and Native Americans. (C and D) Relative cross coalescence rates (CCR) for the Karitiana-Koryak and Athabascan-Koryak divergence (red), respectively, including data simulated under the two demographic models in panel B. In both cases, the model with gene flow (orange) fits the data (red) better than the clean split model (blue). The migration model explains a broader CCR tail in the case of Karitiana-Koryak and the relatively late onset of the CCR decay for Athabascan-Koryak.