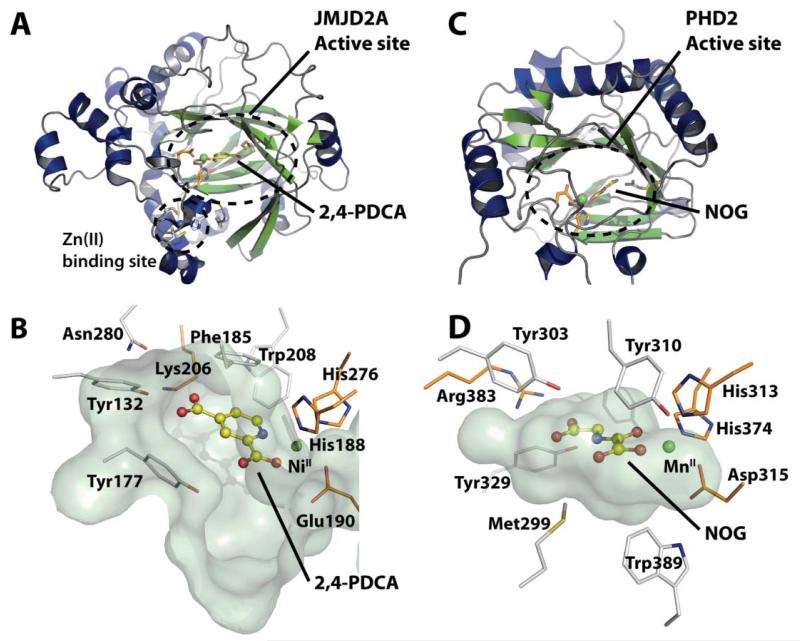
Fig. 1.
Crystallographic analyses on JMJD2A and PHD2. A. View from a crystal structure of JMJD2A in complex with NiII (substituting for FeII) and the inhibitor 2,4-PDCA (derived from PDB ID 2VD715). B. Active site of JMJD2A showing chelation of the active site metal (green) by the conserved iron-binding Hx[D/E] … H motif (His188, Glu190 and His276). Selected active site residues are shown. C. View from a crystal structure of the prolyl hydroxylase PHD2 in complex with MnII, a fragment of the hypoxia inducible factor 1α peptide substrate (not shown), and the 2OG cosubstrate analogue N-oxalyl glycine (NOG), a relatively non-specific 2OG oxygenase inhibitor (PDB ID 3HQR22). D. Closeup view of the PHD2 active site showing chelation of MnII (green) by the conserved iron-binding Hx[D/E] … H motif (His313, Asp315 and His374).