FIG 3.
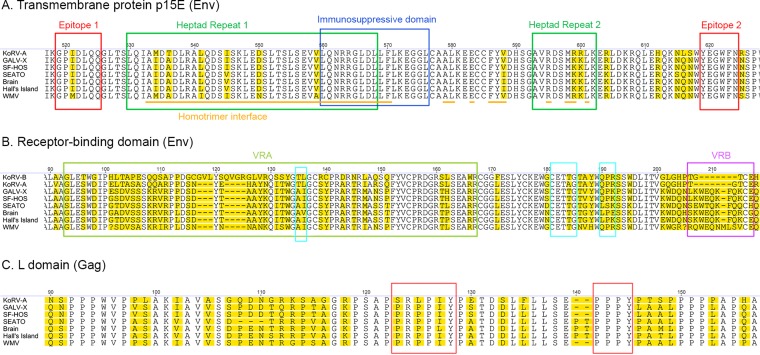
Differences among GALV strains and KoRV in the Env and Gag domains regulating viral fusion, infectivity, and host range. Alignment of Env and Gag amino acid sequences of GALV strains with relevant GenBank reference sequences (GALV-X, U60065; KoRV, AF151794) for the domains affecting viral fusion (epitopes 1 and 2, heptad repeats 1 and 2, homotrimer interface, and immunosuppressive domain of the transmembrane protein p15E of Env) (A), receptor specificity (variable regions A and B of Env) (B), and viral infectivity (receptor-binding domain of Env and L domain of Gag) (B and C, respectively). The three motifs influencing infectivity within the receptor-binding domain are marked by turquoise squares (B), while the PRPPIY and PPPY motifs are marked by brown squares within the L domain (C). Positions where amino acids vary are highlighted in yellow. Since KoRV-B was used to investigate the functional differences between GALV and KoRV in the VRA and VRB regions, KoRV-B has been included in panel B.